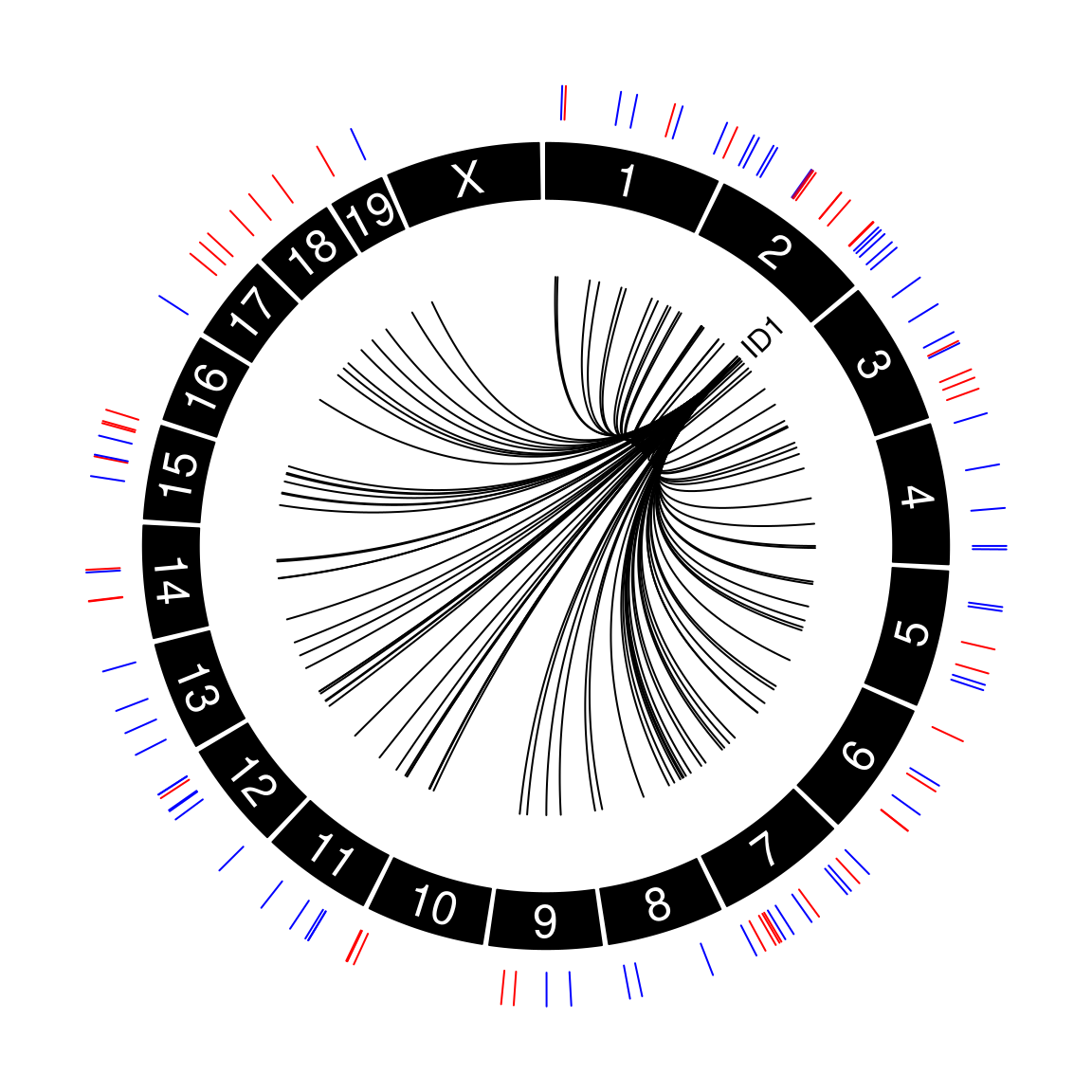
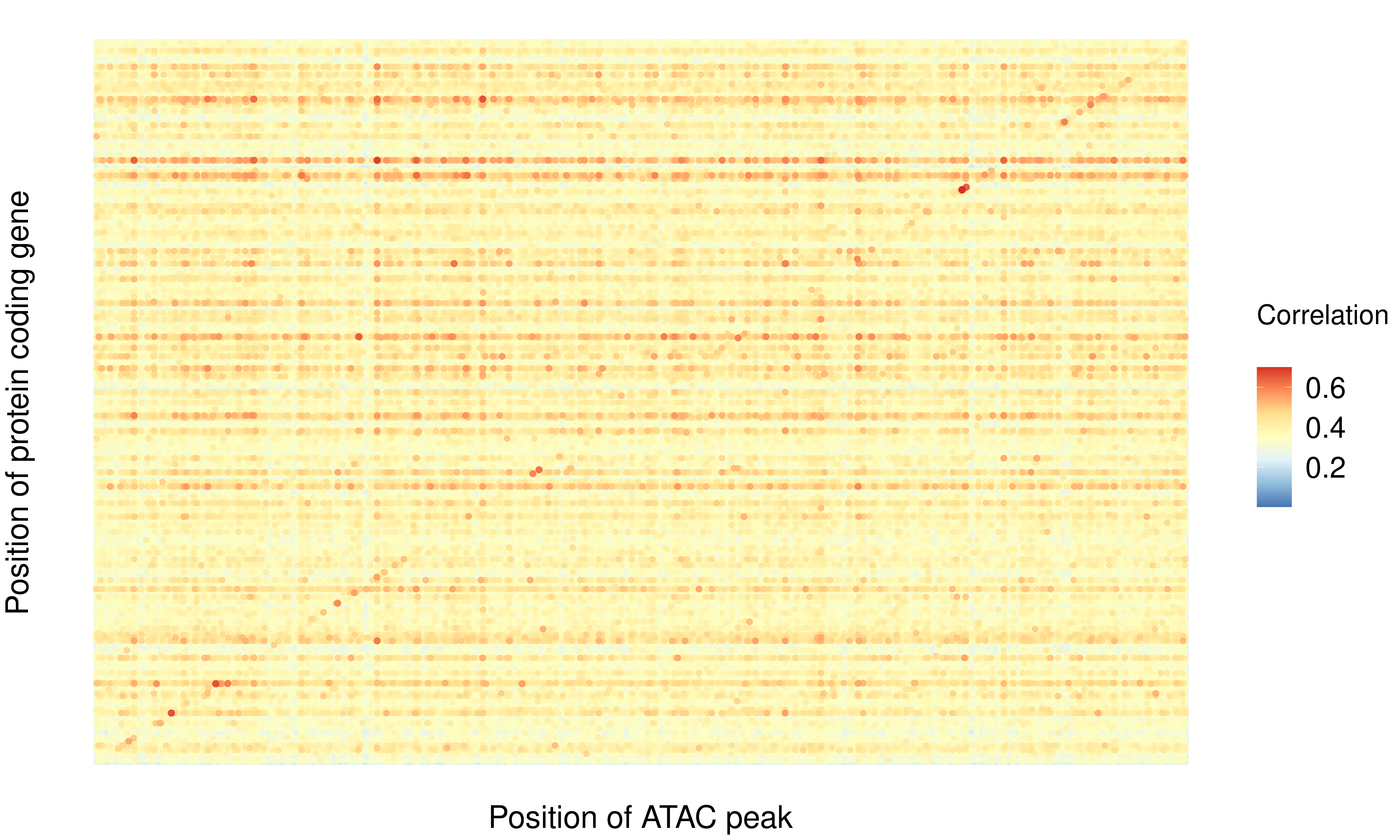
Figure 3A: Covariation of protein abundance and chromatin accessibility
# The code below is not executed because the correlation matrix is very large and takes a long time to run.
# data used to calculate protein vs atac-seq correlations
if (!exists("mm10")) mm10 <- GenomeInfoDb::Seqinfo(genome = "mm10")
snames <- as.character(GenomicRanges::seqnames(mm10)) %>%
grep("^chrUn", ., value = TRUE, invert = TRUE) %>%
grep("_random$", ., value = TRUE, invert = TRUE) %>%
grep("_alt$", ., value = TRUE, invert = TRUE) %>%
grep("_fix$", ., value = TRUE, invert = TRUE) %>%
as_tibble() %>%
filter(!value %in% c("chrX", "chrY", "chrM")) %$% value
autosomes <- mm10[snames]
chrom_lens_Mb <- GenomeInfoDb::seqlengths(mm10)[snames] / 1e6
chrom_lens_Mb_offset <- cumsum(chrom_lens_Mb) - chrom_lens_Mb
binwidth <- 0.5e6
bins <- GenomicRanges::tileGenome(autosomes, tilewidth=binwidth, cut.last.tile.in.chrom=TRUE)
# Some bins (e.g. at end of chroms) are very small
# Merge together any bins that are less than half binwidth with the nearest full-size bin
small_bin_idx <- which(GenomicRanges::width(bins) < binwidth/2)
small_bins <- bins[small_bin_idx]
prev_bins <- bins[small_bin_idx - 1]
#assertthat::assert_that(assertthat::are_equal(GenomicRanges::start(small_bins), GenomicRanges::end(prev_bins)+1))
ii <- setdiff(1:length(bins), c(small_bin_idx, small_bin_idx - 1))
untouched <- bins[ii]
merged <- GenomicRanges::punion(small_bins, prev_bins)
final_bins <- c(untouched, merged) %>% sort()
atac_obj <- tibble(id = rownames(counts.norm2)) %>%
separate(id,
into = c("chrom", "start", "end"),
convert = TRUE, remove = FALSE
) %>%
mutate(chrom = substring(chrom, 5)) %>%
filter(chrom != "Y", chrom != "X", chrom != "MT") %>%
mutate(chrom = paste0("chr", chrom)) %>%
mutate(midpoint = (start + end) / 2) %>%
mutate(start = midpoint, end = midpoint) %>%
GenomicRanges::GRanges(seqinfo = mm10) %>%
sort()
# will be assigning each ATAC peak to one genomic bin based on midpoint
# Find the bin that each ATAC peak falls into
peak_bins <- GenomicRanges::findOverlaps(atac_obj, final_bins)
# assertthat::assert_that(assertthat::are_equal(IRanges::from(peak_bins), 1:length(peak_bins))) # one bin for each peak
prot_dat <- all.prots %>%
mutate(chrom = gene_chr) %>%
select(chrom, gene_start, gene_end, protein_id) %>%
mutate(midpoint=(gene_start+gene_end)/2) %>%
mutate(start=midpoint, end=midpoint) %>%
filter(chrom != "Y", chrom != "X", chrom !="MT") %>%
mutate(chrom=paste0("chr", chrom))
prot_obj <- prot_dat %>%
GenomicRanges::GRanges(seqinfo=mm10) %>% sort()
# Find the bin that each ATAC peak falls into
prot_bins <- GenomicRanges::findOverlaps(prot_obj, final_bins)
# assertthat::assert_that(assertthat::are_equal(IRanges::from(prot_bins), 1:length(prot_bins)))
# Modify expr.both and atac.both to include only autosomes
exprmat <- expr.esc_prot[threeway.shared.samples$sampleid, prot_obj$protein_id]
atacmat <- t(counts.norm2[atac_obj$id, threeway.shared.samples$ATAC])
rownames(atacmat) <- threeway.shared.samples$sampleid
n_eqtl <- length(prot_obj)
n_bins <- length(final_bins)
cors <- cor(exprmat, atacmat, use="pairwise.complete.obs")
cor.prot <- cors # save to variable before getting the abs value for later.
cors <- cors %>% abs()
x <- apply(cors, 1, function(vals)
tapply(vals, INDEX=IRanges::to(peak_bins), FUN=max, na.rm=T))
rownames(x) <- paste0("ATAC_bin", rownames(x))
#low_genes <- apply(x, 2, function(vals) sum(vals > 0.2))
#low_peaks <- apply(x, 1, function(vals) sum(vals > 0.2))
xy <- apply(x, 1, function(vals)
tapply(vals, INDEX=IRanges::to(prot_bins), FUN=mean))
rownames(xy) <- paste0("prot_bin", rownames(xy))
genome_bins_full <- as.data.frame(final_bins) %>% as_tibble() %>%
dplyr::rename(bin_chrom=seqnames) %>%
mutate(bin_midpoint=(start+end)/2) %>%
mutate(bin_width=end-start, full_size=bin_width > binwidth/2) %>%
mutate(n=1:n()) %>%
filter(full_size)
genome_bins <- genome_bins_full %>% select(bin_chrom, bin_midpoint, n)
x2 <- as.data.frame(xy) %>% rownames_to_column("prot_bin") %>%
mutate(prot_bin=substring(prot_bin, 9)) %>%
mutate(prot_bin=as.integer(prot_bin)) %>%
inner_join(genome_bins, by=c("prot_bin"="n")) %>%
dplyr::rename(prot_bin_chrom=bin_chrom, prot_bin_midpoint=bin_midpoint) %>%
gather(ATAC_bin, maxcor, starts_with("ATAC")) %>%
as_tibble() %>% mutate_if(is.factor, as.character) %>%
mutate(ATAC_bin=as.integer(substring(ATAC_bin, 9))) %>%
inner_join(genome_bins, by=c("ATAC_bin"="n")) %>%
dplyr::rename(ATAC_bin_chrom=bin_chrom, ATAC_bin_midpoint=bin_midpoint)
toplot <- select(x2, prot_bin, ATAC_bin, maxcor, prot_bin_chrom ) %>%
dplyr::rename(xpos=ATAC_bin, ypos=prot_bin)
toplot2 <- arrange(toplot, maxcor) %>%
mutate(size_scaled = scales::rescale(maxcor) + 0.1) %>%
mutate(cor_trimmed = ifelse(maxcor > 0.7, 0.7, maxcor)) %>%
mutate(alpha_scaled = scales::rescale(cor_trimmed) + 0.1)
g5.prot <- ggplot(toplot2, aes(xpos, ypos)) +
geom_point(aes(color=cor_trimmed, size=size_scaled, alpha=alpha_scaled)) +
scale_color_distiller(palette="RdYlBu") +
scale_size_continuous(range=c(0.1, 3), guide="none") +
scale_alpha_continuous(range=c(.1, 1), guide="none") +
theme_classic() +
theme(axis.ticks=element_blank(),
axis.text=element_blank(),
axis.line = element_blank(),
axis.title.x = element_text(size = 32),
axis.title.y = element_text(size = 32),
legend.text = element_text(angle=0, hjust =0.1, size = 30),
legend.title = element_text(angle=0, hjust =0, vjust = 0, size = 28)) +
xlab("Position of ATAC peak") +
ylab("Position of protein coding gene") +
coord_cartesian(xlim=c(1, n_bins), ylim=c(1, n_bins)) +
guides(color=guide_colorbar(title="Correlation\n", barwidth=2.5,
barheight=10, bin=100))
ggsave(g5.prot, file = here("figures/mean_sharedProt_maxATAC_05Mb_scaled.png"),width=20, height=12)
# get the max corr protein for each ATACseq peak
(cor.prot)%>%
as_tibble(rownames = "protein_id") %>%
pivot_longer(starts_with("peak"), names_to = "peak_id",values_to="corr") %>%
group_by(peak_id) %>%
slice_max( abs(corr), n = 5) -> max_cor_atac_prot
t(cor.prot)%>%
as_tibble(rownames = "peak_id") %>%
pivot_longer(starts_with("ENS"), names_to = "protein_id",values_to="corr") %>%
filter( abs(corr) > 0.5) %>%
count(protein_id) %>%
left_join(all.prots) -> prot.cor.genes
t(cor.prot)%>%
as_tibble(rownames = "peak_id") %>%
pivot_longer(starts_with("ENS"), names_to = "protein_id",values_to="corr") %>%
filter( abs(corr) > 0.5) %>%
group_by(protein_id) %>%
mutate(n = n()) %>%
filter(n > 100) %>%
ungroup() %>%
left_join(., select(all.prots, "protein_id","cor_gene"="mgi_symbol")) %>%
left_join(atac.peak.annots) -> prot.cor.atacpeaks
save( prot.cor.genes,prot.cor.atacpeaks,max_cor_atac_prot, file = here( "_data/ATAC_prot_cor_data.RData"))load(here("../pQTL_website/_data/ATAC_prot_cor_data.RData"))
prot.cor.atacpeaks %>%
filter(n > 100) %>%
#filter(!cor_gene %in% (filter(rna.cor.genes, n > 50))$mgi_symbol) %>%
select(cor_gene, peak_id, corr) %>%
filter( cor_gene == "Id1") %>%
separate( peak_id, into = c("atac_chr","peak_start","peak_end"), remove = FALSE, sep="_") %>%
mutate( atac_chr = gsub("peak","chr",atac_chr),
peak_start = as.numeric(peak_start),
peak_end = as.numeric(peak_end),
value = 1) %>%
# adding Id1 gene_start and gene_end
left_join(., select(all.genes, mgi_symbol, gene_start,gene_end, gene_chr),
by =c("cor_gene"="mgi_symbol")) %>%
mutate( gene_chr = paste0("chr",gene_chr) ) -> id1_cor_peaks
# atacseq peaks
id1_cor_peaks %>%
select( chr = atac_chr, start = peak_start, end = peak_end, Correlation = corr) %>%
mutate( start = as.integer(start),
end = as.integer(end)
)-> id1_atac_peaks
#id1
id1_cor_peaks %>%
select( chr = gene_chr, start = gene_start, end = gene_end) %>%
mutate( start = as.integer(start),
end = as.integer(end)
) -> id1_genelibrary(circlize)
# to add chromosomes
chroms <- c(as.character(1:19), "X")
chrom_lens <- c( 195431559, 182107670, 160017104, 156496071, 151833620, 149721874, 145434693, 129399468, 124582650, 130685419, 122078650, 120120622 ,120387272, 124867725, 104015452, 98180002, 94984432, 90672596, 61417310 , 171028300)
names(chrom_lens) <- chroms
sectors <- tibble( chr = paste0("chr",chroms),
start = 0,
end = chrom_lens)
circos.par(start.degree = 90, gap.degree= 1)
circos.genomicInitialize(sectors, plotType = NULL)
circos.genomicTrack(id1_atac_peaks,
ylim = c(0,1),
stack = TRUE,
panel.fun = function(region, value, ...) {
i = getI(...)
circos.genomicRect(region, value,
col = ifelse(value[[1]] > 0, "red", "blue") ,
border = ifelse(value[[1]] > 0, "red", "blue"))
}, track.height = 0.1, bg.border = NA)
circos.genomicTrack(stack = TRUE,
ylim = c(1,2),
panel.fun = function(x, y,...) {
i = getI(...)
chr = gsub("chr","",CELL_META$sector.index)
xlim = CELL_META$xlim
ylim = CELL_META$ylim
circos.rect(xlim[1], 0.5, xlim[2], 1.5, col = "black")
circos.text(mean(xlim),1, chr, cex = 1.5, col = "white",
facing = "inside", niceFacing = TRUE)
}, track.height = 0.15, bg.border = NA)
circos.genomicLabels( distinct(id1_gene),
labels = "ID1",
labels.side = "outside",
padding = 0.01,
connection_height = mm_h(0.1),
line_lwd = 0,
cex = 1)
circos.genomicLink(id1_gene,
id1_atac_peaks,
border = "black" )circos.clear()
Data used to generate Figure 3A can be downloaded below.
list(id1_atac_peaks, id1_gene) %>%
downloadthis::download_this(
output_name = "Figure3A data",
output_extension = ".xlsx",
button_label = "Download Figure 3A data as xlsx",
button_type = "primary",
has_icon = TRUE,
icon = "fa fa-save"
)
List of chromatin regions showing high correlation to protein abundance across the genome
prot.cor.atacpeaks %>%
filter(n > 100) %>%
#filter(!cor_gene %in% (filter(rna.cor.genes, n > 50))$mgi_symbol) %>%
select(cor_gene, peak_id, corr, annotation, mgi_symbol, gene_start, gene_end, gene_chr, n) %>%
rename(`# of correalated ATAC-seq peaks` = n) %>%
mutate(corr = formatC(corr, format = "g", digits = 2)) %>%
group_by(peak_id) %>%
mutate(n_gene = n()) %>%
select(cor_gene, peak_id, corr, annotation, mgi_symbol, n_gene, `# of correalated ATAC-seq peaks`) %>%
ungroup() %>%
group_by(cor_gene) %>%
ungroup() %>%
distinct() %>%
mutate( cor_gene = toupper(cor_gene)) %>%
select( `Protein`= (cor_gene),
`Peak id` = peak_id,
`Correlation` = corr,
`Peak annotation (function)` = annotation,
`Peak annotation (gene)` = mgi_symbol,
`# of correalated ATAC-seq peaks`,
`# of correalated proteins` = n_gene
) %>%
create_dt()
Over-representation of transcription binding sites in chromatin regions with high correlation to protein abundance across the genome
background_atac_peaks <- tibble( peak_id = atac.peak.annots_full$peak_id ) %>%
separate( peak_id, into = c("Chr", "Start","End"), remove = FALSE) %>%
mutate( Chr = gsub("peak","chr",Chr)) %>%
makeGRangesFromDataFrame(.,
keep.extra.columns = F,
seqnames.field = c("Chr"),
start.field = "Start",
end.field = "End")
prot.cor.atacpeaks %>%
filter(n > 100) %>%
ungroup() %>%
filter( corr > 0) %>%
left_join(., select(all.prots, "cor_gene_id" = "protein_id", "cor_gene" = "mgi_symbol")) %>%
left_join(atac.peak.annots) %>%
select(cor_gene_id, cor_gene, peak_id, corr, annotation, mgi_symbol) %>%
mutate(corr = formatC(corr, format = "g", digits = 2)) %>%
arrange(cor_gene) %>%
filter(!is.na(mgi_symbol)) %>%
as_tibble() %>%
select(cor_gene, peak_id) %>%
#filter(cor_gene =="Ahdc1") %>%
separate( peak_id, into = c("Chr", "Start","End"), remove = FALSE) %>%
mutate( Chr = gsub("peak","chr",Chr)) %>%
group_by(cor_gene) %>%
nest() %>%
mutate(geneSet = map(data, function(df) {
genes = makeGRangesFromDataFrame( data,
keep.extra.columns = F,
seqnames.field = c("Chr"),
start.field = "Start",
end.field = "End")
return(genes)
})) %>%
select(-data) %>%
ungroup() %>%
mutate( userSet = seq(1:n()), type ="pos")-> all_prot_atac_peaks_pos
prot.cor.atacpeaks %>%
filter(n > 100) %>%
ungroup() %>%
filter( corr < 0) %>%
#filter(!cor_gene %in% (filter(rna.cor.genes, n > 50))$mgi_symbol) %>%
left_join(., select(all.prots, "cor_gene_id" = "protein_id", "cor_gene" = "mgi_symbol")) %>%
left_join(atac.peak.annots) %>%
select(cor_gene_id, cor_gene, peak_id, corr, annotation, mgi_symbol) %>%
mutate(corr = formatC(corr, format = "g", digits = 2)) %>%
arrange(cor_gene) %>%
filter(!is.na(mgi_symbol)) %>%
as_tibble() %>%
select(cor_gene, peak_id) %>%
#filter(cor_gene =="Ahdc1") %>%
separate( peak_id, into = c("Chr", "Start","End"), remove = FALSE) %>%
mutate( Chr = gsub("peak","chr",Chr)) %>%
group_by(cor_gene) %>%
nest() %>%
mutate(geneSet = map(data, function(df) {
genes = makeGRangesFromDataFrame( data,
keep.extra.columns = F,
seqnames.field = c("Chr"),
start.field = "Start",
end.field = "End")
return(genes)
})) %>%
select(-data) %>%
ungroup() %>%
mutate( userSet = seq(1:n()), type ="neg")-> all_prot_atac_peaks_neg
genesSets_pos <- GRangesList(c(all_prot_atac_peaks_pos$geneSet)
)
genesSets_neg <- GRangesList(c(all_prot_atac_peaks_neg$geneSet)
)
ora_prot_unique_atac_peaks_pos <- runLOLA(genesSets_pos,
background_atac_peaks,
regionDB,
cores=1)
ora_prot_unique_atac_peaks_neg <- runLOLA(genesSets_neg,
background_atac_peaks,
regionDB,
cores=1)
ora_prot_unique_atac_peaks_pos$qValue <- (qvalue( 10^(-ora_prot_unique_atac_peaks_pos$pValueLog )))$qvalues
ora_prot_unique_atac_peaks_neg$qValue <- (qvalue( 10^(-ora_prot_unique_atac_peaks_neg$pValueLog )))$qvalues
ora_prot_unique_atac_peaks_neg %>%
left_join( select(all_prot_atac_peaks_neg, userSet, cor_gene) ) %>%
filter( qValue < 0.05) %>%
filter( cellType %in% c("Embryonic Stem Cell", "ES-Bruce4","Embyonic stem cell","Embryonic Stem Cells", "Embryonic stem cells", "embryonic stem cells","embryonic stem cell") ) %>%
group_by(cor_gene ) %>%
mutate( Genes = paste( unique(antibody), collapse = ", "),
n = n_distinct(antibody)) %>%
arrange( desc(n)) %>%
mutate( cor_gene = toupper(cor_gene)) %>%
select( `Protein`= (cor_gene),
`Overrepresented TF binding sites in negatively correlated ATAC-seq peaks` = Genes) %>%
distinct() %>%
full_join(
ora_prot_unique_atac_peaks_pos %>%
left_join( select(all_prot_atac_peaks_pos, userSet, cor_gene) ) %>%
filter( qValue < 0.05) %>%
filter( cellType %in% c("Embryonic Stem Cell", "ES-Bruce4","Embyonic stem cell","Embryonic Stem Cells", "Embryonic stem cells", "embryonic stem cells","embryonic stem cell") ) %>%
group_by( cor_gene) %>%
mutate( Genes = paste( unique(antibody), collapse = ", "),
n = n_distinct(antibody)) %>%
arrange( desc(n)) %>%
mutate(cor_gene = toupper(cor_gene)) %>%
select( `Protein`= (cor_gene),
`Overrepresented TF binding sites in positively correlated ATAC-seq peaks` = Genes) %>%
distinct()
) -> all_lola_results
all_lola_results %>%
create_dt()
Table S4: List of proteins with high correlation to ATAC-seq peaks genome wide.
The table includes additional annotations such as cellular location, Interpro domains, over-represented transcription factor binding sites in ATAC-seq peaks with negative and positive correlations, and relevant references highlighting roles in pluripotency regulation for each protein.
# get the list of proteins that show high correlation (abs(cor)>0.5) to at least 100 ATAC-seq peaks
all_annot_v98_wGO <- read_tsv( here("../pQTL_website/_data","ensembl_gene_annotations_v98_wGO.txt")) %>%
rename( "ensembl_gene_id" = "Gene stable ID",
"protein_id" = "Protein stable ID",
"gene_start" = "Gene start (bp)",
"gene_end" = "Gene end (bp)",
"gene_chr" = "Chromosome/scaffold name",
"mgi_symbol" = "MGI symbol",
"gene_biotype" = "Gene type",
"GO_term_name"="GO term name",
"GO_term_def" = "GO term definition",
"GO_domain" = "GO domain",
"GO_ID"= "GO term accession"
)
all_annot_w98_winterpro <- read_tsv( here("../pQTL_website/_data","ensembl_v98_interpro.txt")) %>%
rename( "ensembl_gene_id" = "Gene stable ID",
"protein_id" = "Protein stable ID") %>%
select(protein_id, interpro_domains =`Interpro Description` ) %>%
distinct()
prot.cor.atacpeaks %>%
filter(n > 100) %>%
ungroup() %>%
mutate( cor_type = ifelse( corr > 0, "Positive", "Negative")) %>%
group_by( cor_gene, cor_type) %>%
count() %>%
pivot_wider( cor_gene, names_from = "cor_type", values_from = "n") %>%
mutate( sum = Positive + Negative) %>%
mutate( `Number of correlated ATAC-seq peaks (positive / negative)` = str_c(sum, " (",Positive,"/", Negative,") ")) %>%
select( cor_gene,`Number of correlated ATAC-seq peaks (positive / negative)`) -> peak_nums
prot.cor.genes %>%
filter(n > 100) %>%
# add cellular location
left_join( all_annot_v98_wGO %>%
filter( GO_domain =="cellular_component") %>%
select( protein_id,
GO_term_name
)
) %>%
group_by(mgi_symbol, protein_id, gene_chr, n) %>%
summarise(across(GO_term_name, str_c, collapse=" ; ")) -> prot_locations
prot.cor.genes %>%
filter(n > 100) %>%
# add interpro domains
left_join( all_annot_w98_winterpro) %>%
group_by(mgi_symbol, protein_id, gene_chr, n) %>%
summarise(across(interpro_domains, str_c, collapse=" ; ")) -> prot_interpro_doms
peak_nums %>%
rename(mgi_symbol = cor_gene) %>%
left_join( prot_locations) %>%
left_join( prot_interpro_doms) %>%
mutate(mgi_symbol = toupper(mgi_symbol),
gene_chr = as.numeric(gene_chr)) %>%
select(`Protein ID` = protein_id,
`Protein` = (mgi_symbol),
`Protein location (Chr)` = gene_chr,
`Cellular location` = GO_term_name,
`Interpro domain` = interpro_domains,
`Number of correlated ATAC-seq peaks (positive / negative)`
) %>%
left_join(
all_lola_results
) %>%
# Ahdc1 annotation is missing, fixing that
mutate( `Cellular location` = ifelse( Protein =="AHDC1", "nucleus", `Cellular location`)) %>%
# add observed at the transcript level?
mutate(`Observed at the transctript level?` = ifelse(
Protein %in% c("ARHGEF1","GJB3","NAPRT", "OOEP", "PHLDA2", "PUS7L"),
"Yes", "No")) %>%
# add references
mutate(
`Published role in pluripotency [ref]` = case_when(
Protein == "AHDC1"~"Moreira S, Seo C, Gordon V, Xing S, Wu R, Polena E, et al. Endogenous BioID elucidates TCF7L1 interactome modulation upon GSK-3 inhibition in mouse ESCs. 2018 Oct p. 431023. doi:10.1101/431023",
Protein =="ID1"~"Romero-Lanman, E.E., Pavlovic, S., Amlani, B., Chin, Y., and Benezra, R. (2012). Id1 Maintains Embryonic Stem Cell Self-Renewal by Up-Regulation of Nanog and Repression of Brachyury Expression. Stem Cells Dev. 21, 384–393.",
Protein =="UHRF2"~"Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010 Feb 5;6(2):153-66. doi: 10.1016/j.stem.2009.12.014. PMID: 20144788; PMCID: PMC2849004."
)) -> table_s4
writexl::write_xlsx( table_s4,
path = here("TableS4_Proteins_w_corr_toATACseq.xlsx"),
col_names = TRUE,
format_headers = TRUE
)
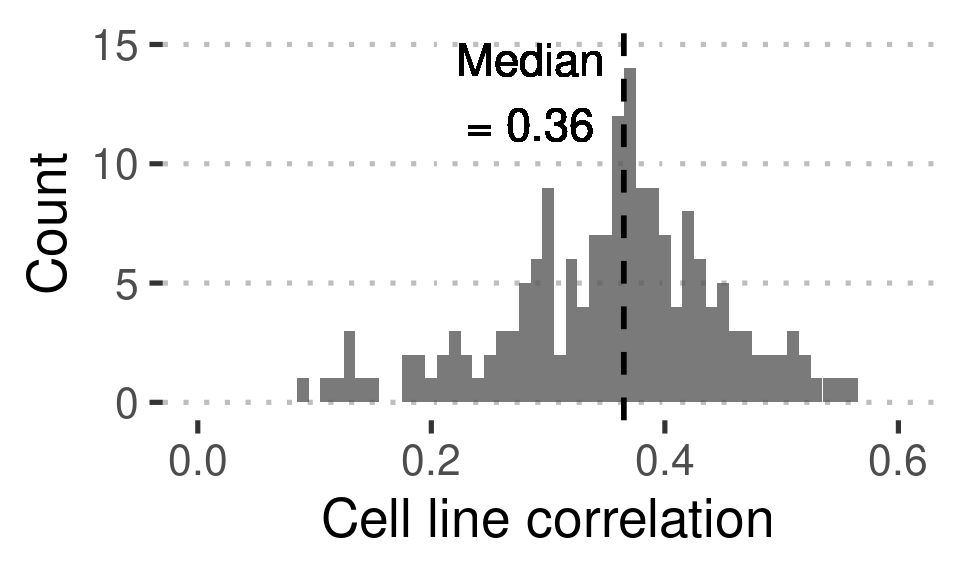
Figure 3B: Covariation of proteome and transcriptome across samples
shared.prot.names <- shared.genes %>%
group_by(ensembl_gene_id) %>%
mutate(new_symbol = paste0(mgi_symbol, "_", 1:n()),
new_gene_id = paste0(ensembl_gene_id, "_", 1:n()))
shared_prot_mat <- t(exprZ.esc_prot[shared.samples, shared.prot.names$protein_id])
colnames(shared_prot_mat) <- paste0(colnames(shared_prot_mat),"_protein")
shared_rna_mat <- t(exprZ.esc_rna[shared.samples, shared.prot.names$ensembl_gene_id])
colnames(shared_rna_mat) <- paste0(colnames(shared_rna_mat),"_rna")
prot_rna_sample_cor <- rcorr( x = shared_prot_mat,
y = shared_rna_mat,
type = "pearson")
prot_rna_sample_cor_df <- as_tibble( prot_rna_sample_cor$r[colnames(shared_prot_mat), colnames(shared_rna_mat)],
rownames = "protein_sample") %>%
pivot_longer( colnames(shared_rna_mat), names_to = "rna_sample", values_to = "r") %>%
inner_join( (as_tibble( prot_rna_sample_cor$P[colnames(shared_prot_mat), colnames(shared_rna_mat)],
rownames = "protein_sample") %>%
pivot_longer( colnames(shared_rna_mat), names_to = "rna_sample", values_to = "p_val") ) )
prot_rna_sample_cor_df %>%
mutate( sampleid_prot = gsub("_protein","",protein_sample),
sampleid_rna = gsub("_rna","", rna_sample)) %>%
filter( sampleid_rna == sampleid_prot) %>% summarize( med_r = median(r)) -> median_cor_rna_prot
Figure3B_data <- prot_rna_sample_cor_df %>%
mutate( sampleid_prot = gsub("_protein","",protein_sample),
sampleid_rna = gsub("_rna","", rna_sample)) %>%
filter( sampleid_rna == sampleid_prot) %>%
rename( `Sample id` = sampleid_prot,
`Correlation` = r)Figure3B_data %>%
ggplot() +
aes(x = Correlation) +
geom_histogram( show.legend = F, binwidth = 0.01, alpha = 0.8) +
geom_vline( aes(xintercept = median(Correlation,na.rm=T)), color = "black", linetype = "dashed", size = 1 )+
geom_text( mapping= aes(
x = median(Correlation,na.rm=T)-0.08,
label = stringr::str_wrap(paste0("Median = ",round(median(Correlation,na.rm=T),2)),7),
),
y = 13,
size = 6
)+
xlab("Cell line correlation") +
ylab("Count") +
theme_pubclean(base_size = 20)+
xlim(0,0.6)+
ylim(0,15)
Figure3B_data %>%
select(
`Sample id` ,`Correlation`
) %>%
mutate_if(is.numeric, round ,2) %>%
create_dt()
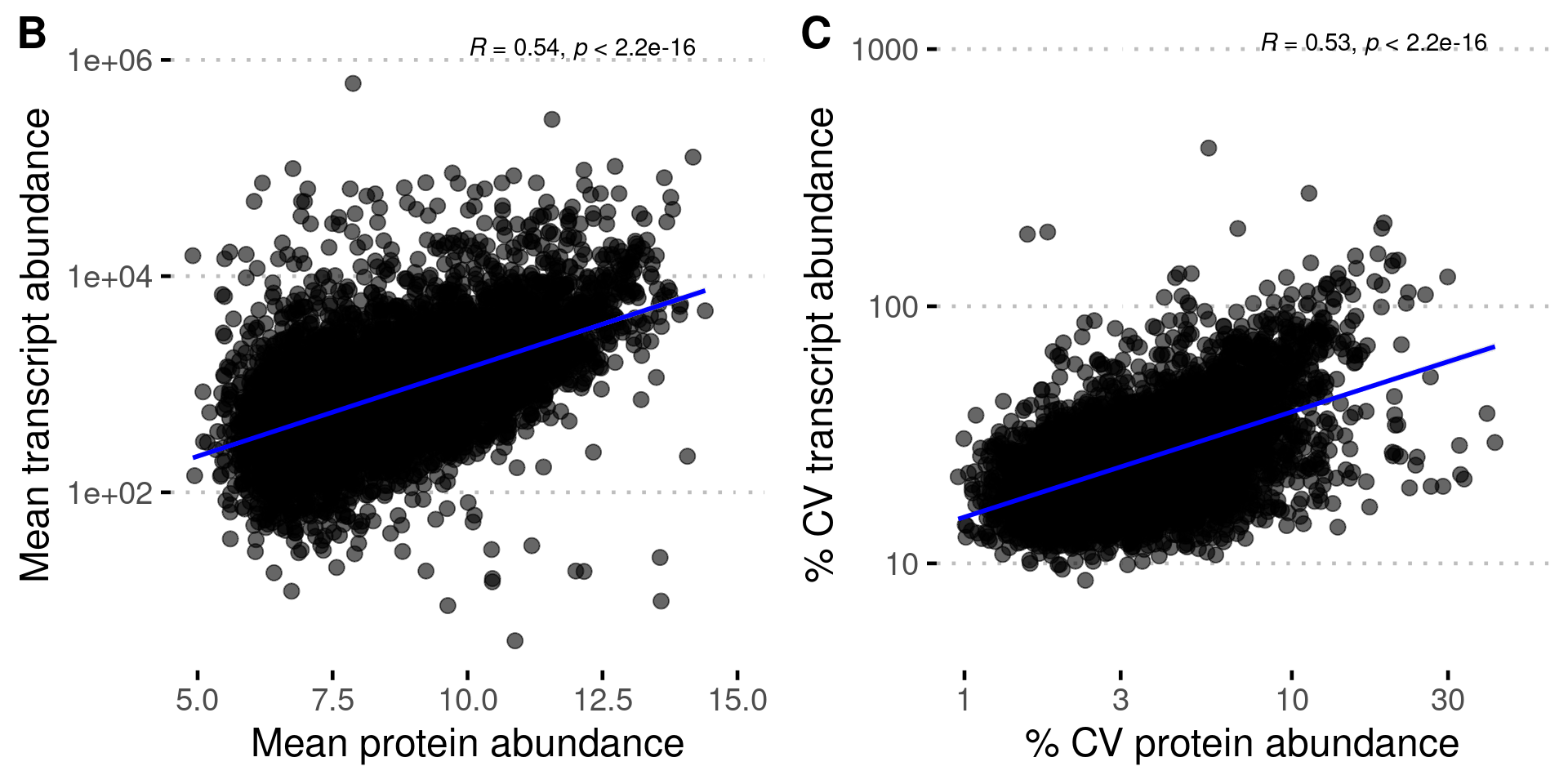
Figure S3B-C: Variation in transcript and protein abundance
# get mean + %cv for protein abundance
var_prot <- expr.esc_prot %>%
as_tibble(.) %>%
summarise_all(list(~ var(., na.rm = T))) %>%
pivot_longer(everything(),
names_to = "protein_id",
values_to ="var.prot")
mean_prot <- expr.esc_prot %>%
as_tibble(.) %>%
summarise_all(list(~ mean(., na.rm = T))) %>%
pivot_longer( everything(),
names_to = "protein_id",
values_to ="mean.prot")
var_prot %>%
full_join( mean_prot) %>%
mutate(sd.prot = sqrt(var.prot)) %>%
mutate(cv.prot = 100 * sd.prot / (mean.prot)) -> all_var_prot
# get mean + %cv for transcript abundance
var_rna <- expr.esc_rna %>%
as_tibble(.) %>%
summarise_all(list(~ var(., na.rm = T))) %>%
pivot_longer( everything(),
names_to = "ensembl_gene_id",
values_to ="var.rna")
mean_rna <- expr.esc_rna %>%
as_tibble(.) %>%
summarise_all(list(~ mean(., na.rm = T))) %>%
pivot_longer( everything(),
names_to = "ensembl_gene_id",
values_to ="mean.rna")
var_rna %>%
full_join( mean_rna) %>%
left_join( all.genes) %>%
mutate(sd.rna = sqrt(var.rna)) %>%
mutate(cv.rna = 100 * sd.rna / (mean.rna)) -> all_var_rna
# join transcript + protein variation
all_var_rna %>%
left_join( shared.genes) %>%
inner_join(all_var_prot %>%
left_join(shared.genes)) -> all_var
# plotting figures S3b-c
all_var %>%
ggscatter(
.,
x = "mean.prot", y = "mean.rna", size = 3, alpha = 0.6,
add = "reg.line", # Add regression line
conf.int = TRUE, # Add confidence interval
add.params = list(color = "blue", fill = "lightgray"), show.legend.text = FALSE,
yscale = "log10"
) +
stat_cor(method = "pearson", label.x = 10, label.y = 6.1) + # Add correlation coefficient
# stat_cor( aes(label = paste(..rr.label.., ..p.label.., sep = "~`,`~")),
# label.x = 0.65, label.y = 6) +# Add regression R2
xlab("Mean protein abundance") +
ylab("Mean transcript abundance") +
theme_pubclean(base_size = 18) + rremove("legend") -> figure_s3b
figure_s3b <- ggpar(figure_s3b, xlim = c(5, 15))
all_var %>%
ggscatter(. ,
x = "cv.prot", y = "cv.rna", size = 3, alpha = 0.6,
add = "reg.line", # Add regression line
conf.int = TRUE, # Add confidence interval
add.params = list(color = "blue", fill = "lightgray"), show.legend.text = FALSE,
yscale = "log10",
xscale = "log10"
) +
stat_cor(method = "pearson", label.x = 0.9, label.y = 3.02) + # Add correlation coefficient
# stat_cor( aes(label = paste(..rr.label.., ..p.label.., sep = "~`,`~")),
# label.x = -0.01, label.y = 3) +# Add regression R2
xlab("% CV protein abundance") +
ylab("% CV transcript abundance") +
theme_pubclean(base_size = 18) + rremove("legend") -> figure_s3c
figure_s3c <- ggpar( figure_s3c, xlim= c(1, 50), ylim = c(5, 1000))
ggarrange(figure_s3b,
figure_s3c,
nrow = 1,
labels = c("B","C"),
font.label = list( size = 20)
)
Genes with difference in variation in transcript and protein abundance
# genes with high variation in protein abundance and low variation in transcript abundance
all_var %>%
filter( !is.na(cv.rna), !is.na(cv.prot)) %>%
filter( cv.prot > quantile(cv.prot, 0.75) & cv.rna < quantile(cv.rna, 0.25) ) %>%
left_join(all.prots) -> rev_var_prots
# genes with high variation in transcript abundance and low variation in protein abundance
all_var %>%
filter( !is.na(cv.rna), !is.na(cv.prot)) %>%
filter( cv.prot < quantile(cv.prot, 0.25) & cv.rna > quantile(cv.rna, 0.75) ) %>%
left_join(all.prots) -> rev_var_prots2
ora_rev_var_prots <- gost( query = unique(rev_var_prots$mgi_symbol),
organism = "mmusculus",
domain_scope = "custom",
custom_bg = shared.genes$mgi_symbol,
evcodes = TRUE,
correction_method = "fdr"
)
ora_rev_var_prots$result <- filter( ora_rev_var_prots$result, term_size < 600) # nothing over-represented, empty!
ora_rev_var_prots2 <- gost( query = unique(rev_var_prots2$mgi_symbol),
organism = "mmusculus",
domain_scope = "custom",
custom_bg = shared.genes$mgi_symbol,
evcodes = TRUE,
correction_method = "fdr"
)
ora_rev_var_prots2$result <- filter( ora_rev_var_prots2$result, term_size < 600)
ora_rev_var_prots2$result %>%
select(
`Data source` = source,
`Term ID` = term_id,
`Term Name` = term_name,
`Term size` = term_size,
`# of intersecting proteins` = intersection_size,
FDR = p_value
) %>%
mutate_if( is.numeric, formatC, digits =2) %>%
create_dt()
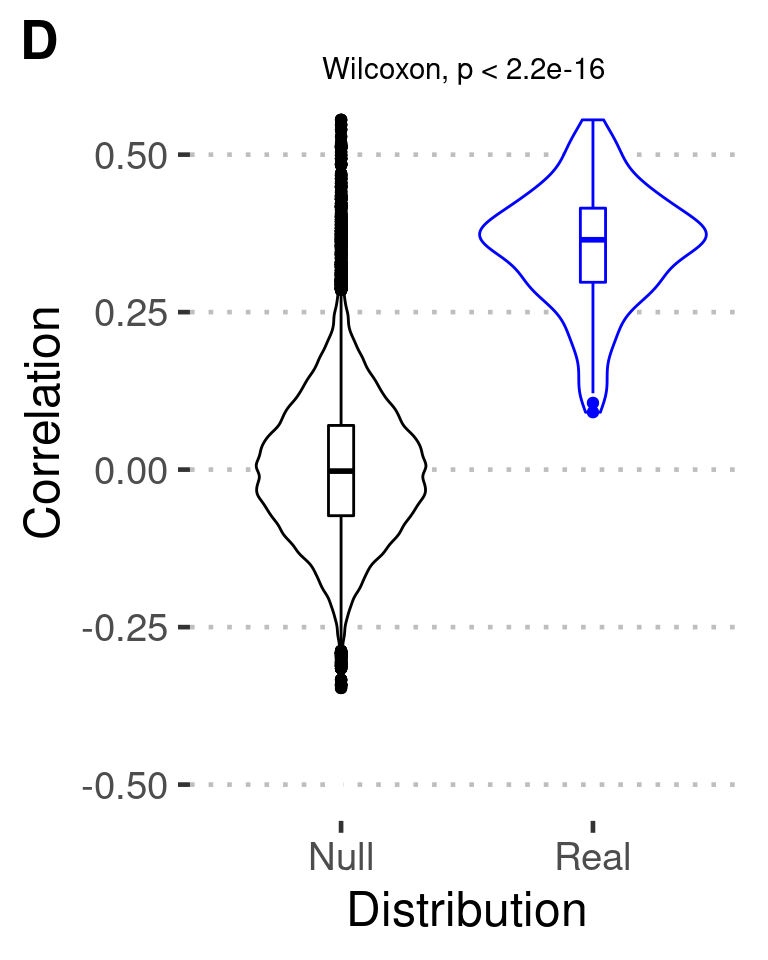
Figure S3D
# The code below is used to generate the null distribution.
# It is commented out and loaded from the saved Rdata file because it takes too long to run within the script.
# sample_cor <- c()
# for( i in 1:1000){
# # randomizing the sample names 1000 times and getting correlations
#
# shared_prot_mat <- t(exprZ.esc_prot[shared.samples, shared.prot.names$protein_id])
# # randomize the sample names
# colnames(shared_prot_mat) <- paste0( sample(colnames(shared_prot_mat), ncol(shared_prot_mat)),"_protein")
#
# shared_rna_mat <- t(exprZ.esc_rna[shared.samples, shared.prot.names$ensembl_gene_id])
# # randomize the sample names
# colnames(shared_rna_mat) <- paste0( sample(colnames(shared_rna_mat), ncol(shared_rna_mat)),"_rna")
#
# measure.cor.df <- rcorr( x = shared_prot_mat,
# y = shared_rna_mat,
# type = "pearson")
#
# sample_cor[[i]] <- as_tibble( measure.cor.df$r[colnames(shared_prot_mat), colnames(shared_rna_mat)],
# rownames = "protein_sample") %>%
# pivot_longer( colnames(shared_rna_mat), names_to = "rna_sample", values_to = "r") %>%
# inner_join( (as_tibble( measure.cor.df$P[colnames(shared_prot_mat), colnames(shared_rna_mat)],
# rownames = "protein_sample") %>%
# pivot_longer( colnames(shared_rna_mat), names_to = "rna_sample", values_to = "p_val") ) ) %>%
# mutate( n = i)
#
# }
#
# save(sample_cor, file = here("_data","rna_prot_sample_cor_perm_pearson.RData"))
load(here("../pQTL_website/_data","rna_prot_sample_cor_perm_pearson.RData"))
sample_cor %>%
enframe() %>%
unnest(value) %>%
mutate( protein_sample = gsub("_protein","",protein_sample) ,
rna_sample = gsub("_rna","",rna_sample)) %>%
filter( protein_sample == rna_sample) -> null_sample_cor_dist
prot_rna_sample_cor_df %>%
mutate( sampleid_prot = gsub("_protein","",protein_sample),
sampleid_rna = gsub("_rna","", rna_sample)) %>%
filter( sampleid_rna == sampleid_prot) -> real_sample_cor_dist
null_sample_cor_dist %>%
mutate( type = "Null") %>%
select( type, r) %>%
rbind( real_sample_cor_dist %>%
mutate( type = "Real") %>%
select( type, r)) %>%
ggplot()+
aes( x = type,
y = r,
col = type )+
geom_violin( show.legend = F)+
geom_boxplot(width = 0.1, show.legend = F)+
scale_color_manual( values = c("black","blue"))+
theme_pubclean(base_size = 18)+
theme(legend.position="none")+
ylab("Correlation")+
xlab("Distribution")+
stat_compare_means( label.y = 0.62, label.x = 1.15)+
ylim(-0.5,.65) -> fig_s3d
ggarrange(fig_s3d,
labels = "D",
font.label = list( size = 20)
)
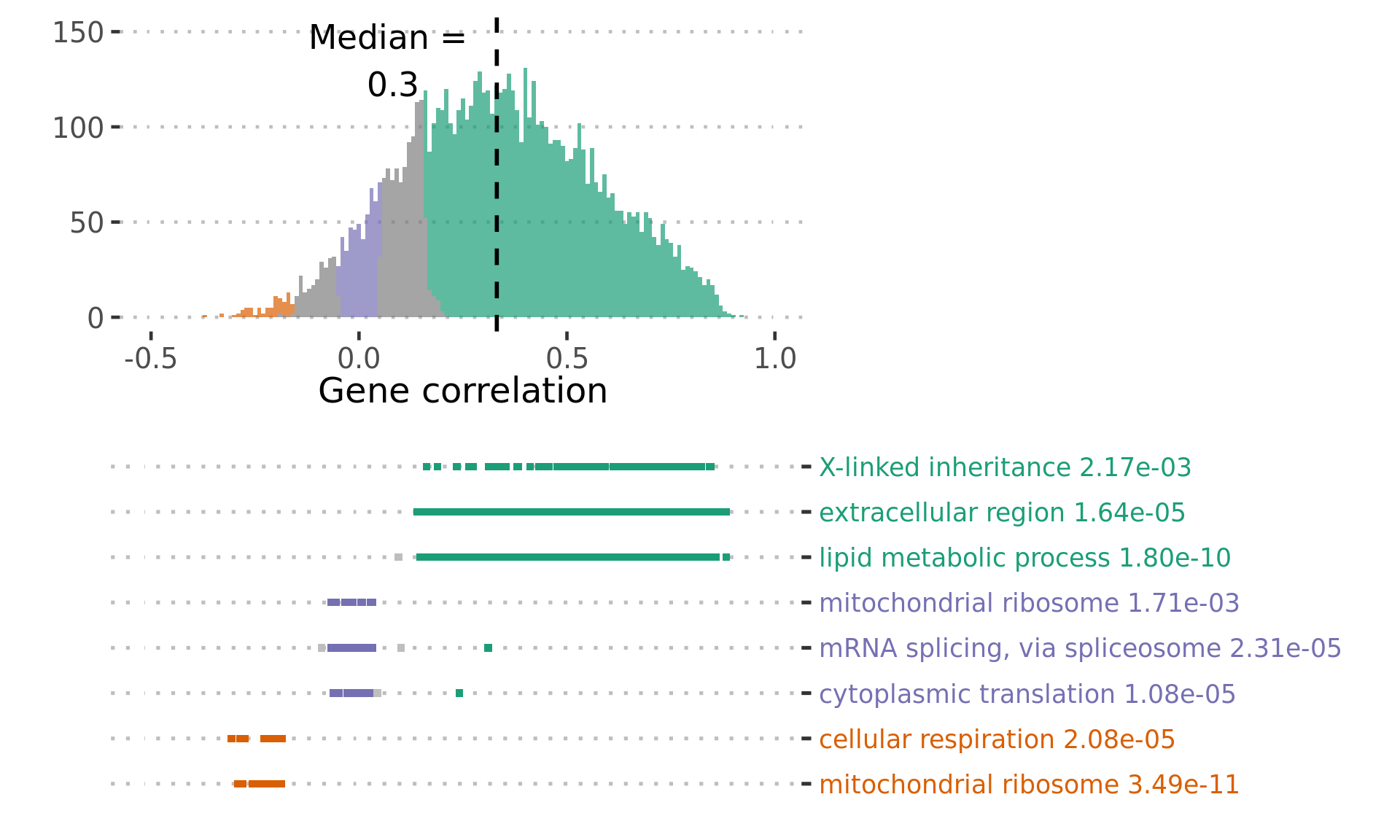
Figure 3C: Covariation of protein and transcript abundance across genes
# get gene names for all protein ids
shared.prot.names <- shared.genes %>%
group_by(ensembl_gene_id) %>%
mutate(new_symbol = paste0(mgi_symbol, "_", 1:n()),
new_gene_id = paste0(ensembl_gene_id, "_", 1:n()))
# change id's into gene symbols
shared_prot_mat2 <- (exprZ.esc_prot[shared.samples, shared.prot.names$protein_id])
shared_rna_mat2 <- (exprZ.esc_rna[shared.samples, shared.prot.names$ensembl_gene_id])
# get gene level correlations
prot_rna_gene_cor <- rcorr( x = shared_prot_mat2,
y = shared_rna_mat2,
type = "pearson")
# convert gene level correlations to data frame
prot_rna_gene_cor_df <- tibble( r =diag(prot_rna_gene_cor$r[colnames(shared_prot_mat2), colnames(shared_rna_mat2)]),
p_val = diag(prot_rna_gene_cor$P[colnames(shared_prot_mat2), colnames(shared_rna_mat2)]),
n = diag(prot_rna_gene_cor$n[colnames(shared_prot_mat2), colnames(shared_rna_mat2)]),
protein_id = colnames(shared_prot_mat2),
ensembl_gene_id = colnames(shared_rna_mat2)
) %>%
left_join(., shared.prot.names) %>%
mutate(p_adj = p.adjust(p_val, method = "BH")) %>%
mutate( cor = r) %>%
arrange((desc(cor))) %>%
mutate(rank = seq(1:n()))
# get significantly negatively and positively correlated genes and genes with not correlation (abs(cor) <0.05).
neg_cor <- prot_rna_gene_cor_df %>%
filter( cor < 0, p_adj < 0.05)
pos_cor <- prot_rna_gene_cor_df %>%
filter( cor > 0, p_adj < 0.05) %>%
arrange( desc(cor) )
no_cor <- prot_rna_gene_cor_df %>%
filter( abs(cor) < 0.05)
# over-representation analysis with each category
ora_neg_cor <- gost( query = neg_cor$mgi_symbol,
organism = "mmusculus",
domain_scope = "custom",
custom_bg = prot_rna_gene_cor_df$mgi_symbol,
evcodes = TRUE,
correction_method = "fdr"
)
ora_neg_cor$result <- filter( ora_neg_cor$result, term_size < 600)
ora_pos_cor <- gost( query = pos_cor$mgi_symbol,
organism = "mmusculus",
domain_scope = "custom",
custom_bg = prot_rna_gene_cor_df$mgi_symbol,
evcodes = TRUE,
correction_method = "fdr"
)
ora_pos_cor$result <- filter( ora_pos_cor$result, term_size < 600)
ora_no_cor <- gost( query = no_cor$mgi_symbol,
organism = "mmusculus",
domain_scope = "custom",
custom_bg = prot_rna_gene_cor_df$mgi_symbol,
evcodes = TRUE,
correction_method = "fdr"
)
ora_no_cor$result <- filter( ora_no_cor$result, term_size < 600)
# merge all ORA results into one data frame
ora_neg_cor$result %>%
mutate( group = "Negative correlation") %>%
rbind( mutate( ora_pos_cor$result, group = "Positive correlation")) %>%
rbind( mutate( ora_no_cor$result , group = "No correlation")) -> ora_all_corr
Figure3C_data_part1 <- prot_rna_gene_cor_df %>%
mutate(
Group = case_when(
(cor < 0 & p_adj < 0.05) ~ "Negative",
(cor > 0 & p_adj < 0.05) ~ "Positive",
( abs(cor) < 0.05) ~ "Low"
)
) %>%
select(
`Gene name` = mgi_symbol,
`Protein ID` = protein_id,
`Gene ID` = ensembl_gene_id,
`Correlation` = cor,
Group
)
# get these:
# negative: cellular respiration, mitochondrial ribosome
# no correlation: Ribosome, Spliceosome OR cytoplasmic translation, mRNA splicing, via spliceosome
# positive correlation: extracellular region, lipid metabolic process
# rank here is the order above
# For adding the dots below the correlation histogram
Figure3C_data_part2 <- ora_all_corr %>%
#filter(source %in% c("GO:BP","REAC","KEGG")) %>%
filter(term_name %in% c("cellular respiration", "mitochondrial ribosome",
"cytoplasmic translation", "mRNA splicing, via spliceosome",
"extracellular region", "lipid metabolic process","X-linked inheritance") ) %>%
select(term_name, intersection, p_value, group) %>%
arrange(desc(p_value), group) %>%
separate_rows(intersection) %>%
left_join(., prot_rna_gene_cor_df, by = c("intersection" = "mgi_symbol")) %>%
mutate( group2 = case_when( ( cor < 0 & p_adj < 0.05) ~ "negative",
( cor > 0 & p_adj < 0.05) ~ "positive",
( abs(cor) <= 0.05 ) ~ "low",
( abs(cor) > 0.05 & p_adj >= 0.05) ~ "none"
)
)
y_colors <- RColorBrewer::brewer.pal(n = 4, name = "Dark2")
# For adding the labels next to dots in matching color
Figure3C_data_part3 <- ora_all_corr %>%
filter(term_name %in% c("cellular respiration", "mitochondrial ribosome",
"cytoplasmic translation", "mRNA splicing, via spliceosome",
"extracellular region", "lipid metabolic process","X-linked inheritance") ) %>%
select(term_name, intersection, p_value, group) %>%
mutate(term_name = paste(term_name, formatC(p_value, format = "e", digits = 2))) %>%
separate_rows(intersection) %>%
left_join(., prot_rna_gene_cor_df, by = c("intersection" = "mgi_symbol")) %>%
group_by(term_name, group, p_value) %>%
dplyr::summarize(med_cor = median(cor, na.rm = TRUE)) %>%
mutate(y_col = ifelse(group == "Negative correlation", y_colors[2],y_colors[4]),
y_col = ifelse(group == "No correlation", y_colors[3], y_col),
y_col = ifelse(group == "Positive correlation", y_colors[1], y_col)) %>%
arrange(group, p_value) # plot results
y_colors <- RColorBrewer::brewer.pal(n = 4, name = "Dark2")
Figure3C_data_part1 %>%
ggplot() +
aes(x = Correlation) +
geom_histogram(aes(fill = Group), show.legend = F, binwidth = 0.01, alpha = 0.7) +
xlab("Gene correlation") +
ylab("")+
scale_fill_manual(limits = c( "Low",NA, "Negative", "Positive"), values = c(y_colors[3],"gray",y_colors[2],y_colors[1]) ) +
theme_pubclean(base_size = 18, base_family = "Poppins")+
geom_vline( aes(xintercept = median(Correlation)), linetype = 2, col = "black", size = 1) +
annotate("text",
x = median(Figure3C_data_part1$Correlation, na.rm = T) -0.25, y = 135,
label = paste0("Median = \n", formatC(median(Figure3C_data_part1$Correlation, na.rm = T), digits = 1, format ="f")),
size = 6,
family = "Poppins"
)+
xlim(-0.5, 1)+
ylim(0,150)-> p1
Figure3C_data_part2 %>%
arrange(cor) %>%
mutate(label2 = factor(paste(term_name, formatC(p_value, format = "e", digits = 2)),
levels = Figure3C_data_part3$term_name
)) %>%
mutate(label = factor(intersection, levels = unique(intersection))) %>%
ggplot() +
aes(y = label2, x = cor, col = group2, group = term_name) +
geom_point(show.legend = FALSE, shape = 15) +
scale_color_manual(limits = c("low", "none", "negative", "positive"), values = c(y_colors[3],"gray",y_colors[2],y_colors[1]) ) +
xlab("Gene correlation") +
ylab("") +
theme_pubclean(base_size = 20, base_family = "Poppins") +
theme(
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.y = element_text(size = 13, color = (Figure3C_data_part3$y_col))
) +
scale_y_discrete(position = "right") +
xlim(-0.5, 1)-> p2
top_row <- plot_grid(p1, NULL, rel_widths = c(1, 0.7))
bottom_row <- plot_grid(NULL, p2, rel_widths = c(0.07, 1))
ora_cor_plot<- plot_grid(top_row, bottom_row, nrow = 2, rel_heights = c(1, 1))
ora_cor_plot
Figure3C_data_part1 %>%
mutate_if( is.numeric, round, 2) %>%
create_dt()
ora_all_corr %>%
select(
`Group`=group,
`Data source` = source,
`Term ID` = term_id,
`Term Name` = term_name,
`Term size` = term_size,
`# of intersecting proteins` = intersection_size,
FDR = p_value
) %>%
mutate_if( is.numeric, formatC, digits =2) %>%
create_dt()
LS0tCnRpdGxlOiAiQ28tdmFyaWF0aW9uIGluIHByb3RlaW4gYWJ1bmRhbmNlLCBjaHJvbWF0aW4gYWNjZXNzaWJpbGl0eSBhbmQgdHJhbnNjcmlwdCBhYnVuZGFuY2UiCm91dHB1dDoKICBodG1sX2RvY3VtZW50OgogICAgdG9jOiB0cnVlCiAgICB0b2NfZGVwdGg6IDQKICAgIHRvY19mbG9hdDogCiAgICAgIGNvbGxhcHNlZDogZmFsc2UKICAgICAgc21vb3RoX3Njcm9sbDogZmFsc2UKICAgIGRmX3ByaW50OiBwYWdlZAogICAgY29kZV9mb2xkaW5nOiBoaWRlCi0tLQoKPHN0eWxlPgpwLmNhcHRpb24gewogIGZvbnQtc2l6ZTogMWVtOwp9Cjwvc3R5bGU+CgoKYGBge3Igc2V0dXB9CgojIG9wdGlvbnMKb3B0aW9ucyhzdHJpbmdzQXNGYWN0b3JzID0gRikKa25pdHI6Om9wdHNfY2h1bmskc2V0KGVjaG8gPSBUUlVFLCB3YXJuaW5nID0gRkFMU0UsIG1lc3NhZ2UgPSBGQUxTRSkKa25pdHI6Om9wdHNfa25pdCRzZXQocHJvZ3Jlc3MgPSBGQUxTRSkKCmBgYAoKPGJyPgo8YnI+CgojIyMgRmlndXJlIDNBOiBDb3ZhcmlhdGlvbiBvZiBwcm90ZWluIGFidW5kYW5jZSBhbmQgY2hyb21hdGluIGFjY2Vzc2liaWxpdHkKCgpgYGB7ciBQcm90ZWluX2F0YWNfU2VxX2NvciwgZXZhbCA9IEZBTFNFfQojIFRoZSBjb2RlIGJlbG93IGlzIG5vdCBleGVjdXRlZCBiZWNhdXNlIHRoZSBjb3JyZWxhdGlvbiBtYXRyaXggaXMgdmVyeSBsYXJnZSBhbmQgdGFrZXMgYSBsb25nIHRpbWUgdG8gcnVuLgoKIyBkYXRhIHVzZWQgdG8gY2FsY3VsYXRlIHByb3RlaW4gdnMgYXRhYy1zZXEgY29ycmVsYXRpb25zCmlmICghZXhpc3RzKCJtbTEwIikpIG1tMTAgPC0gR2Vub21lSW5mb0RiOjpTZXFpbmZvKGdlbm9tZSA9ICJtbTEwIikKc25hbWVzIDwtIGFzLmNoYXJhY3RlcihHZW5vbWljUmFuZ2VzOjpzZXFuYW1lcyhtbTEwKSkgJT4lCiAgZ3JlcCgiXmNoclVuIiwgLiwgdmFsdWUgPSBUUlVFLCBpbnZlcnQgPSBUUlVFKSAlPiUKICBncmVwKCJfcmFuZG9tJCIsIC4sIHZhbHVlID0gVFJVRSwgaW52ZXJ0ID0gVFJVRSkgJT4lCiAgZ3JlcCgiX2FsdCQiLCAuLCB2YWx1ZSA9IFRSVUUsIGludmVydCA9IFRSVUUpICU+JQogIGdyZXAoIl9maXgkIiwgLiwgdmFsdWUgPSBUUlVFLCBpbnZlcnQgPSBUUlVFKSAlPiUKICBhc190aWJibGUoKSAlPiUKICBmaWx0ZXIoIXZhbHVlICVpbiUgYygiY2hyWCIsICJjaHJZIiwgImNock0iKSkgJSQlIHZhbHVlCmF1dG9zb21lcyA8LSBtbTEwW3NuYW1lc10KY2hyb21fbGVuc19NYiA8LSBHZW5vbWVJbmZvRGI6OnNlcWxlbmd0aHMobW0xMClbc25hbWVzXSAvIDFlNgpjaHJvbV9sZW5zX01iX29mZnNldCA8LSBjdW1zdW0oY2hyb21fbGVuc19NYikgLSBjaHJvbV9sZW5zX01iCgpiaW53aWR0aCA8LSAwLjVlNgpiaW5zIDwtIEdlbm9taWNSYW5nZXM6OnRpbGVHZW5vbWUoYXV0b3NvbWVzLCB0aWxld2lkdGg9Ymlud2lkdGgsIGN1dC5sYXN0LnRpbGUuaW4uY2hyb209VFJVRSkKIyBTb21lIGJpbnMgKGUuZy4gYXQgZW5kIG9mIGNocm9tcykgYXJlIHZlcnkgc21hbGwKIyBNZXJnZSB0b2dldGhlciBhbnkgYmlucyB0aGF0IGFyZSBsZXNzIHRoYW4gaGFsZiBiaW53aWR0aCB3aXRoIHRoZSBuZWFyZXN0IGZ1bGwtc2l6ZSBiaW4Kc21hbGxfYmluX2lkeCA8LSB3aGljaChHZW5vbWljUmFuZ2VzOjp3aWR0aChiaW5zKSA8IGJpbndpZHRoLzIpCnNtYWxsX2JpbnMgPC0gYmluc1tzbWFsbF9iaW5faWR4XQpwcmV2X2JpbnMgPC0gYmluc1tzbWFsbF9iaW5faWR4IC0gMV0KI2Fzc2VydHRoYXQ6OmFzc2VydF90aGF0KGFzc2VydHRoYXQ6OmFyZV9lcXVhbChHZW5vbWljUmFuZ2VzOjpzdGFydChzbWFsbF9iaW5zKSwgR2Vub21pY1Jhbmdlczo6ZW5kKHByZXZfYmlucykrMSkpCmlpIDwtIHNldGRpZmYoMTpsZW5ndGgoYmlucyksIGMoc21hbGxfYmluX2lkeCwgc21hbGxfYmluX2lkeCAtIDEpKQp1bnRvdWNoZWQgPC0gYmluc1tpaV0KbWVyZ2VkIDwtIEdlbm9taWNSYW5nZXM6OnB1bmlvbihzbWFsbF9iaW5zLCBwcmV2X2JpbnMpCmZpbmFsX2JpbnMgPC0gYyh1bnRvdWNoZWQsIG1lcmdlZCkgJT4lIHNvcnQoKQoKYXRhY19vYmogPC0gdGliYmxlKGlkID0gcm93bmFtZXMoY291bnRzLm5vcm0yKSkgJT4lCiAgc2VwYXJhdGUoaWQsCiAgICBpbnRvID0gYygiY2hyb20iLCAic3RhcnQiLCAiZW5kIiksCiAgICBjb252ZXJ0ID0gVFJVRSwgcmVtb3ZlID0gRkFMU0UKICApICU+JQogIG11dGF0ZShjaHJvbSA9IHN1YnN0cmluZyhjaHJvbSwgNSkpICU+JQogIGZpbHRlcihjaHJvbSAhPSAiWSIsIGNocm9tICE9ICJYIiwgY2hyb20gIT0gIk1UIikgJT4lCiAgbXV0YXRlKGNocm9tID0gcGFzdGUwKCJjaHIiLCBjaHJvbSkpICU+JQogIG11dGF0ZShtaWRwb2ludCA9IChzdGFydCArIGVuZCkgLyAyKSAlPiUKICBtdXRhdGUoc3RhcnQgPSBtaWRwb2ludCwgZW5kID0gbWlkcG9pbnQpICU+JQogIEdlbm9taWNSYW5nZXM6OkdSYW5nZXMoc2VxaW5mbyA9IG1tMTApICU+JQogIHNvcnQoKQojIHdpbGwgYmUgYXNzaWduaW5nIGVhY2ggQVRBQyBwZWFrIHRvIG9uZSBnZW5vbWljIGJpbiBiYXNlZCBvbiBtaWRwb2ludAojIEZpbmQgdGhlIGJpbiB0aGF0IGVhY2ggQVRBQyBwZWFrIGZhbGxzIGludG8KcGVha19iaW5zIDwtIEdlbm9taWNSYW5nZXM6OmZpbmRPdmVybGFwcyhhdGFjX29iaiwgZmluYWxfYmlucykKIyBhc3NlcnR0aGF0Ojphc3NlcnRfdGhhdChhc3NlcnR0aGF0OjphcmVfZXF1YWwoSVJhbmdlczo6ZnJvbShwZWFrX2JpbnMpLCAxOmxlbmd0aChwZWFrX2JpbnMpKSkgIyBvbmUgYmluIGZvciBlYWNoIHBlYWsKCgpwcm90X2RhdCA8LSBhbGwucHJvdHMgJT4lCiAgIG11dGF0ZShjaHJvbSA9IGdlbmVfY2hyKSAlPiUKICAgIHNlbGVjdChjaHJvbSwgZ2VuZV9zdGFydCwgZ2VuZV9lbmQsIHByb3RlaW5faWQpICU+JQogICAgbXV0YXRlKG1pZHBvaW50PShnZW5lX3N0YXJ0K2dlbmVfZW5kKS8yKSAlPiUgCiAgICBtdXRhdGUoc3RhcnQ9bWlkcG9pbnQsIGVuZD1taWRwb2ludCkgJT4lCiAgICBmaWx0ZXIoY2hyb20gIT0gIlkiLCBjaHJvbSAhPSAiWCIsIGNocm9tICE9Ik1UIikgJT4lIAogICAgbXV0YXRlKGNocm9tPXBhc3RlMCgiY2hyIiwgY2hyb20pKSAKcHJvdF9vYmogPC0gcHJvdF9kYXQgJT4lCiAgR2Vub21pY1Jhbmdlczo6R1JhbmdlcyhzZXFpbmZvPW1tMTApICU+JSBzb3J0KCkKCiMgRmluZCB0aGUgYmluIHRoYXQgZWFjaCBBVEFDIHBlYWsgZmFsbHMgaW50bwpwcm90X2JpbnMgPC0gR2Vub21pY1Jhbmdlczo6ZmluZE92ZXJsYXBzKHByb3Rfb2JqLCBmaW5hbF9iaW5zKQojIGFzc2VydHRoYXQ6OmFzc2VydF90aGF0KGFzc2VydHRoYXQ6OmFyZV9lcXVhbChJUmFuZ2VzOjpmcm9tKHByb3RfYmlucyksIDE6bGVuZ3RoKHByb3RfYmlucykpKQoKIyBNb2RpZnkgZXhwci5ib3RoIGFuZCBhdGFjLmJvdGggdG8gaW5jbHVkZSBvbmx5IGF1dG9zb21lcwpleHBybWF0IDwtIGV4cHIuZXNjX3Byb3RbdGhyZWV3YXkuc2hhcmVkLnNhbXBsZXMkc2FtcGxlaWQsIHByb3Rfb2JqJHByb3RlaW5faWRdCmF0YWNtYXQgPC0gdChjb3VudHMubm9ybTJbYXRhY19vYmokaWQsIHRocmVld2F5LnNoYXJlZC5zYW1wbGVzJEFUQUNdKQpyb3duYW1lcyhhdGFjbWF0KSA8LSB0aHJlZXdheS5zaGFyZWQuc2FtcGxlcyRzYW1wbGVpZAoKbl9lcXRsIDwtIGxlbmd0aChwcm90X29iaikKbl9iaW5zIDwtIGxlbmd0aChmaW5hbF9iaW5zKQpjb3JzIDwtIGNvcihleHBybWF0LCBhdGFjbWF0LCB1c2U9InBhaXJ3aXNlLmNvbXBsZXRlLm9icyIpIApjb3IucHJvdCA8LSBjb3JzICMgc2F2ZSB0byB2YXJpYWJsZSBiZWZvcmUgZ2V0dGluZyB0aGUgYWJzIHZhbHVlIGZvciBsYXRlci4KY29ycyA8LSBjb3JzICU+JSBhYnMoKQp4IDwtIGFwcGx5KGNvcnMsIDEsIGZ1bmN0aW9uKHZhbHMpCiAgICB0YXBwbHkodmFscywgSU5ERVg9SVJhbmdlczo6dG8ocGVha19iaW5zKSwgRlVOPW1heCwgbmEucm09VCkpCnJvd25hbWVzKHgpIDwtIHBhc3RlMCgiQVRBQ19iaW4iLCByb3duYW1lcyh4KSkKCiNsb3dfZ2VuZXMgPC0gYXBwbHkoeCwgMiwgZnVuY3Rpb24odmFscykgc3VtKHZhbHMgPiAwLjIpKQojbG93X3BlYWtzIDwtIGFwcGx5KHgsIDEsIGZ1bmN0aW9uKHZhbHMpIHN1bSh2YWxzID4gMC4yKSkKeHkgPC0gYXBwbHkoeCwgMSwgZnVuY3Rpb24odmFscykKICB0YXBwbHkodmFscywgSU5ERVg9SVJhbmdlczo6dG8ocHJvdF9iaW5zKSwgRlVOPW1lYW4pKQpyb3duYW1lcyh4eSkgPC0gcGFzdGUwKCJwcm90X2JpbiIsIHJvd25hbWVzKHh5KSkKZ2Vub21lX2JpbnNfZnVsbCA8LSBhcy5kYXRhLmZyYW1lKGZpbmFsX2JpbnMpICU+JSBhc190aWJibGUoKSAlPiUKICBkcGx5cjo6cmVuYW1lKGJpbl9jaHJvbT1zZXFuYW1lcykgJT4lCiAgbXV0YXRlKGJpbl9taWRwb2ludD0oc3RhcnQrZW5kKS8yKSAlPiUKICBtdXRhdGUoYmluX3dpZHRoPWVuZC1zdGFydCwgZnVsbF9zaXplPWJpbl93aWR0aCA+IGJpbndpZHRoLzIpICU+JQogIG11dGF0ZShuPTE6bigpKSAlPiUKICBmaWx0ZXIoZnVsbF9zaXplKQpnZW5vbWVfYmlucyA8LSBnZW5vbWVfYmluc19mdWxsICU+JSBzZWxlY3QoYmluX2Nocm9tLCBiaW5fbWlkcG9pbnQsIG4pCngyIDwtIGFzLmRhdGEuZnJhbWUoeHkpICU+JSByb3duYW1lc190b19jb2x1bW4oInByb3RfYmluIikgJT4lCiAgbXV0YXRlKHByb3RfYmluPXN1YnN0cmluZyhwcm90X2JpbiwgOSkpICU+JQogIG11dGF0ZShwcm90X2Jpbj1hcy5pbnRlZ2VyKHByb3RfYmluKSkgJT4lCiAgaW5uZXJfam9pbihnZW5vbWVfYmlucywgYnk9YygicHJvdF9iaW4iPSJuIikpICU+JQogIGRwbHlyOjpyZW5hbWUocHJvdF9iaW5fY2hyb209YmluX2Nocm9tLCBwcm90X2Jpbl9taWRwb2ludD1iaW5fbWlkcG9pbnQpICU+JQogIGdhdGhlcihBVEFDX2JpbiwgbWF4Y29yLCBzdGFydHNfd2l0aCgiQVRBQyIpKSAlPiUKICBhc190aWJibGUoKSAlPiUgbXV0YXRlX2lmKGlzLmZhY3RvciwgYXMuY2hhcmFjdGVyKSAlPiUKICBtdXRhdGUoQVRBQ19iaW49YXMuaW50ZWdlcihzdWJzdHJpbmcoQVRBQ19iaW4sIDkpKSkgJT4lCiAgaW5uZXJfam9pbihnZW5vbWVfYmlucywgYnk9YygiQVRBQ19iaW4iPSJuIikpICU+JQogIGRwbHlyOjpyZW5hbWUoQVRBQ19iaW5fY2hyb209YmluX2Nocm9tLCBBVEFDX2Jpbl9taWRwb2ludD1iaW5fbWlkcG9pbnQpCgp0b3Bsb3QgPC0gc2VsZWN0KHgyLCBwcm90X2JpbiwgQVRBQ19iaW4sIG1heGNvciwgcHJvdF9iaW5fY2hyb20gKSAlPiUKICBkcGx5cjo6cmVuYW1lKHhwb3M9QVRBQ19iaW4sIHlwb3M9cHJvdF9iaW4pIAp0b3Bsb3QyIDwtIGFycmFuZ2UodG9wbG90LCBtYXhjb3IpICU+JQogIG11dGF0ZShzaXplX3NjYWxlZCA9IHNjYWxlczo6cmVzY2FsZShtYXhjb3IpICsgMC4xKSAlPiUKICBtdXRhdGUoY29yX3RyaW1tZWQgPSBpZmVsc2UobWF4Y29yID4gMC43LCAwLjcsIG1heGNvcikpICU+JQogIG11dGF0ZShhbHBoYV9zY2FsZWQgPSBzY2FsZXM6OnJlc2NhbGUoY29yX3RyaW1tZWQpICsgMC4xKQpnNS5wcm90IDwtIGdncGxvdCh0b3Bsb3QyLCBhZXMoeHBvcywgeXBvcykpICsKICBnZW9tX3BvaW50KGFlcyhjb2xvcj1jb3JfdHJpbW1lZCwgc2l6ZT1zaXplX3NjYWxlZCwgYWxwaGE9YWxwaGFfc2NhbGVkKSkgKwogIHNjYWxlX2NvbG9yX2Rpc3RpbGxlcihwYWxldHRlPSJSZFlsQnUiKSArCiAgc2NhbGVfc2l6ZV9jb250aW51b3VzKHJhbmdlPWMoMC4xLCAzKSwgZ3VpZGU9Im5vbmUiKSArCiAgc2NhbGVfYWxwaGFfY29udGludW91cyhyYW5nZT1jKC4xLCAxKSwgZ3VpZGU9Im5vbmUiKSArCiAgdGhlbWVfY2xhc3NpYygpICsKICB0aGVtZShheGlzLnRpY2tzPWVsZW1lbnRfYmxhbmsoKSwgCiAgICAgICAgYXhpcy50ZXh0PWVsZW1lbnRfYmxhbmsoKSwgCiAgICAgICAgYXhpcy5saW5lID0gZWxlbWVudF9ibGFuaygpLAogICAgICAgIGF4aXMudGl0bGUueCA9IGVsZW1lbnRfdGV4dChzaXplID0gMzIpLAogICAgICAgIGF4aXMudGl0bGUueSA9IGVsZW1lbnRfdGV4dChzaXplID0gMzIpLAogICAgICAgIGxlZ2VuZC50ZXh0ID0gZWxlbWVudF90ZXh0KGFuZ2xlPTAsIGhqdXN0ID0wLjEsIHNpemUgPSAzMCksCiAgICAgICAgbGVnZW5kLnRpdGxlID0gZWxlbWVudF90ZXh0KGFuZ2xlPTAsIGhqdXN0ID0wLCB2anVzdCA9IDAsIHNpemUgPSAyOCkpICsKICB4bGFiKCJQb3NpdGlvbiBvZiBBVEFDIHBlYWsiKSArCiAgeWxhYigiUG9zaXRpb24gb2YgcHJvdGVpbiBjb2RpbmcgZ2VuZSIpICsKICBjb29yZF9jYXJ0ZXNpYW4oeGxpbT1jKDEsIG5fYmlucyksIHlsaW09YygxLCBuX2JpbnMpKSArCiAgZ3VpZGVzKGNvbG9yPWd1aWRlX2NvbG9yYmFyKHRpdGxlPSJDb3JyZWxhdGlvblxuIiwgYmFyd2lkdGg9Mi41LAogICAgYmFyaGVpZ2h0PTEwLCBiaW49MTAwKSkKZ2dzYXZlKGc1LnByb3QsIGZpbGUgPSBoZXJlKCJmaWd1cmVzL21lYW5fc2hhcmVkUHJvdF9tYXhBVEFDXzA1TWJfc2NhbGVkLnBuZyIpLHdpZHRoPTIwLCBoZWlnaHQ9MTIpCgoKIyBnZXQgdGhlIG1heCBjb3JyIHByb3RlaW4gZm9yIGVhY2ggQVRBQ3NlcSBwZWFrCihjb3IucHJvdCklPiUKICBhc190aWJibGUocm93bmFtZXMgPSAicHJvdGVpbl9pZCIpICU+JQogIHBpdm90X2xvbmdlcihzdGFydHNfd2l0aCgicGVhayIpLCBuYW1lc190byA9ICJwZWFrX2lkIix2YWx1ZXNfdG89ImNvcnIiKSAlPiUKICBncm91cF9ieShwZWFrX2lkKSAlPiUgCiAgc2xpY2VfbWF4KCBhYnMoY29yciksIG4gPSA1KSAtPiBtYXhfY29yX2F0YWNfcHJvdCAKCnQoY29yLnByb3QpJT4lCiAgYXNfdGliYmxlKHJvd25hbWVzID0gInBlYWtfaWQiKSAlPiUgCiAgcGl2b3RfbG9uZ2VyKHN0YXJ0c193aXRoKCJFTlMiKSwgbmFtZXNfdG8gPSAicHJvdGVpbl9pZCIsdmFsdWVzX3RvPSJjb3JyIikgJT4lCiAgZmlsdGVyKCBhYnMoY29ycikgPiAwLjUpICAlPiUKICBjb3VudChwcm90ZWluX2lkKSAlPiUKICBsZWZ0X2pvaW4oYWxsLnByb3RzKSAtPiBwcm90LmNvci5nZW5lcwoKdChjb3IucHJvdCklPiUKICBhc190aWJibGUocm93bmFtZXMgPSAicGVha19pZCIpICU+JSAKICBwaXZvdF9sb25nZXIoc3RhcnRzX3dpdGgoIkVOUyIpLCBuYW1lc190byA9ICJwcm90ZWluX2lkIix2YWx1ZXNfdG89ImNvcnIiKSAlPiUKICBmaWx0ZXIoIGFicyhjb3JyKSA+IDAuNSkgICU+JQogIGdyb3VwX2J5KHByb3RlaW5faWQpICU+JQogIG11dGF0ZShuID0gbigpKSAlPiUKICBmaWx0ZXIobiA+IDEwMCkgJT4lCiAgdW5ncm91cCgpICU+JQogIGxlZnRfam9pbiguLCBzZWxlY3QoYWxsLnByb3RzLCAicHJvdGVpbl9pZCIsImNvcl9nZW5lIj0ibWdpX3N5bWJvbCIpKSAlPiUKICBsZWZ0X2pvaW4oYXRhYy5wZWFrLmFubm90cykgLT4gcHJvdC5jb3IuYXRhY3BlYWtzCgpzYXZlKCBwcm90LmNvci5nZW5lcyxwcm90LmNvci5hdGFjcGVha3MsbWF4X2Nvcl9hdGFjX3Byb3QsIGZpbGUgPSBoZXJlKCAiX2RhdGEvQVRBQ19wcm90X2Nvcl9kYXRhLlJEYXRhIikpCgoKYGBgCgpgYGB7ciBGaWd1cmUzQV9wcmVwfQoKbG9hZChoZXJlKCIuLi9wUVRMX3dlYnNpdGUvX2RhdGEvQVRBQ19wcm90X2Nvcl9kYXRhLlJEYXRhIikpCgpwcm90LmNvci5hdGFjcGVha3MgJT4lCiAgZmlsdGVyKG4gPiAxMDApICU+JQogICNmaWx0ZXIoIWNvcl9nZW5lICVpbiUgKGZpbHRlcihybmEuY29yLmdlbmVzLCBuID4gNTApKSRtZ2lfc3ltYm9sKSAlPiUKICBzZWxlY3QoY29yX2dlbmUsIHBlYWtfaWQsIGNvcnIpICU+JQogIGZpbHRlciggY29yX2dlbmUgPT0gIklkMSIpICU+JQogIHNlcGFyYXRlKCBwZWFrX2lkLCBpbnRvID0gYygiYXRhY19jaHIiLCJwZWFrX3N0YXJ0IiwicGVha19lbmQiKSwgcmVtb3ZlID0gRkFMU0UsIHNlcD0iXyIpICU+JSAKICBtdXRhdGUoIGF0YWNfY2hyID0gZ3N1YigicGVhayIsImNociIsYXRhY19jaHIpLAogICAgICAgICAgcGVha19zdGFydCA9IGFzLm51bWVyaWMocGVha19zdGFydCksCiAgICAgICAgICBwZWFrX2VuZCA9IGFzLm51bWVyaWMocGVha19lbmQpLAogICAgICAgICAgdmFsdWUgPSAxKSAlPiUgCiAgIyBhZGRpbmcgSWQxIGdlbmVfc3RhcnQgYW5kIGdlbmVfZW5kCiAgbGVmdF9qb2luKC4sIHNlbGVjdChhbGwuZ2VuZXMsIG1naV9zeW1ib2wsIGdlbmVfc3RhcnQsZ2VuZV9lbmQsIGdlbmVfY2hyKSwgCiAgICAgICAgICAgIGJ5ID1jKCJjb3JfZ2VuZSI9Im1naV9zeW1ib2wiKSkgJT4lIAogIG11dGF0ZSggZ2VuZV9jaHIgPSBwYXN0ZTAoImNociIsZ2VuZV9jaHIpICkgLT4gaWQxX2Nvcl9wZWFrcwoKIyBhdGFjc2VxIHBlYWtzCmlkMV9jb3JfcGVha3MgJT4lICAKICBzZWxlY3QoIGNociA9IGF0YWNfY2hyLCBzdGFydCA9IHBlYWtfc3RhcnQsIGVuZCA9IHBlYWtfZW5kLCBDb3JyZWxhdGlvbiA9IGNvcnIpICU+JSAKICBtdXRhdGUoIHN0YXJ0ID0gYXMuaW50ZWdlcihzdGFydCksCiAgICAgICAgICBlbmQgPSBhcy5pbnRlZ2VyKGVuZCkKICAgICAgICAgICktPiBpZDFfYXRhY19wZWFrcwojaWQxCmlkMV9jb3JfcGVha3MgJT4lICAKICBzZWxlY3QoIGNociA9IGdlbmVfY2hyLCBzdGFydCA9IGdlbmVfc3RhcnQsIGVuZCA9IGdlbmVfZW5kKSAlPiUgCiAgbXV0YXRlKCBzdGFydCA9IGFzLmludGVnZXIoc3RhcnQpLAogICAgICAgICAgZW5kID0gYXMuaW50ZWdlcihlbmQpCiAgICAgICAgICApICAtPiBpZDFfZ2VuZQoKYGBgCgoKYGBge3IgRmlndXJlM0FfcGxvdCwgZmlnLmNhcD0iRmlndXJlIDNBOiBJRDEgcHJvdGVpbiBhYnVuZGFuY2Ugc2hvd3MgaGlnaCBjb3JyZWxhdGlvbiB0byBtYW55IGNocm9tYXRpbiByZWdpb25zIGFjcm9zcyB0aGUgZ2Vub21lLiBDaXJjb3MgcGxvdCBzaG93aW5nIEFUQUMtc2VxIHBlYWtzIHdoZXJlIGNocm9tYXRpbiBhY2Nlc3NpYmlsaXR5IGlzIHBvc2l0aXZlbHkgKHJlZCkgYW5kIG5lZ2F0aXZlbHkgKGJsdWUpIGNvcnJlbGF0ZWQgd2l0aCBJRDEgcHJvdGVpbiBhYnVuZGFuY2UgKG4gPSAxMTIsIGFicyhjb3JyZWxhdGlvbikgPjAuNSkuIiwgZmlnLmhlaWdodD02LCBmaWcud2lkdGg9Nn0KCmxpYnJhcnkoY2lyY2xpemUpCgojIHRvIGFkZCBjaHJvbW9zb21lcwpjaHJvbXMgPC0gYyhhcy5jaGFyYWN0ZXIoMToxOSksICJYIikKY2hyb21fbGVucyA8LSBjKCAxOTU0MzE1NTksIDE4MjEwNzY3MCwgMTYwMDE3MTA0LCAxNTY0OTYwNzEsIDE1MTgzMzYyMCwgMTQ5NzIxODc0LCAxNDU0MzQ2OTMsIDEyOTM5OTQ2OCwgMTI0NTgyNjUwLCAxMzA2ODU0MTksIDEyMjA3ODY1MCwgMTIwMTIwNjIyICwxMjAzODcyNzIsIDEyNDg2NzcyNSwgMTA0MDE1NDUyLCA5ODE4MDAwMiwgOTQ5ODQ0MzIsIDkwNjcyNTk2LCA2MTQxNzMxMCAsIDE3MTAyODMwMCkKbmFtZXMoY2hyb21fbGVucykgPC0gY2hyb21zCnNlY3RvcnMgPC0gdGliYmxlKCBjaHIgPSBwYXN0ZTAoImNociIsY2hyb21zKSwKICAgICAgICAgICAgICAgICAgIHN0YXJ0ID0gMCwKICAgICAgICAgICAgICAgICAgIGVuZCA9IGNocm9tX2xlbnMpCgpjaXJjb3MucGFyKHN0YXJ0LmRlZ3JlZSA9IDkwLCBnYXAuZGVncmVlPSAxKQpjaXJjb3MuZ2Vub21pY0luaXRpYWxpemUoc2VjdG9ycywgcGxvdFR5cGUgPSBOVUxMKQpjaXJjb3MuZ2Vub21pY1RyYWNrKGlkMV9hdGFjX3BlYWtzLAogICAgICAgICAgICAgICAgICAgIHlsaW0gPSBjKDAsMSksCiAgICAgICAgICAgICAgICAgICAgc3RhY2sgPSBUUlVFLAogICAgcGFuZWwuZnVuID0gZnVuY3Rpb24ocmVnaW9uLCB2YWx1ZSwgLi4uKSB7CiAgICAgIGkgPSBnZXRJKC4uLikKICAgICAgY2lyY29zLmdlbm9taWNSZWN0KHJlZ2lvbiwgdmFsdWUsCiAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbCA9IGlmZWxzZSh2YWx1ZVtbMV1dID4gMCwgInJlZCIsICJibHVlIikgLAogICAgICAgICAgICAgICAgICAgICAgICAgICBib3JkZXIgPSBpZmVsc2UodmFsdWVbWzFdXSA+IDAsICJyZWQiLCAiYmx1ZSIpKQp9LCB0cmFjay5oZWlnaHQgPSAwLjEsIGJnLmJvcmRlciA9IE5BKQpjaXJjb3MuZ2Vub21pY1RyYWNrKHN0YWNrID0gVFJVRSwKICAgICAgICAgICAgICAgICAgICB5bGltID0gYygxLDIpLAogICAgICAgICAgICAgICAgICAgIHBhbmVsLmZ1biA9IGZ1bmN0aW9uKHgsIHksLi4uKSB7CiAgICAgICAgICAgICAgICAgICAgICBpID0gZ2V0SSguLi4pCiAgICAgICAgICAgICAgICAgICAgICBjaHIgPSBnc3ViKCJjaHIiLCIiLENFTExfTUVUQSRzZWN0b3IuaW5kZXgpCiAgICAgICAgICAgICAgICAgICAgICB4bGltID0gQ0VMTF9NRVRBJHhsaW0KICAgICAgICAgICAgICAgICAgICAgIHlsaW0gPSBDRUxMX01FVEEkeWxpbQogICAgICAgICAgICAgICAgICAgICAgY2lyY29zLnJlY3QoeGxpbVsxXSwgMC41LCB4bGltWzJdLCAxLjUsIGNvbCA9ICJibGFjayIpCiAgICAgICAgICAgICAgICAgICAgICBjaXJjb3MudGV4dChtZWFuKHhsaW0pLDEsIGNociwgY2V4ID0gMS41LCBjb2wgPSAid2hpdGUiLAogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgZmFjaW5nID0gImluc2lkZSIsIG5pY2VGYWNpbmcgPSBUUlVFKQp9LCB0cmFjay5oZWlnaHQgPSAwLjE1LCBiZy5ib3JkZXIgPSBOQSkKY2lyY29zLmdlbm9taWNMYWJlbHMoIGRpc3RpbmN0KGlkMV9nZW5lKSwKICAgICAgICAgICAgICAgICAgICAgIGxhYmVscyA9ICJJRDEiLCAKICAgICAgICAgICAgICAgICAgICAgIGxhYmVscy5zaWRlID0gIm91dHNpZGUiLCAKICAgICAgICAgICAgICAgICAgICAgIHBhZGRpbmcgPSAwLjAxLCAKICAgICAgICAgICAgICAgICAgICAgIGNvbm5lY3Rpb25faGVpZ2h0ID0gbW1faCgwLjEpLAogICAgICAgICAgICAgICAgICAgICAgbGluZV9sd2QgPSAwLAogICAgICAgICAgICAgICAgICAgICAgY2V4ID0gMSkKY2lyY29zLmdlbm9taWNMaW5rKGlkMV9nZW5lLAogICAgICAgICAgICAgICAgICAgaWQxX2F0YWNfcGVha3MsCiAgICAgICAgICAgICAgICAgICBib3JkZXIgPSAiYmxhY2siICkKY2lyY29zLmNsZWFyKCkKCmBgYAoKPGJyPgoKRGF0YSB1c2VkIHRvIGdlbmVyYXRlIEZpZ3VyZSAzQSBjYW4gYmUgZG93bmxvYWRlZCBiZWxvdy4KCmBgYHtyIEZpZ3VyZV8zQV9kYXRhLCBmaWcuY2FwID0gIkRhdGEgdXNlZCBpbiBGaWd1cmUgM0EgdG8gZ2VuZXJhdGUgdGhlIGdlbm9taWMgcmVnaW9ucyBpbiB0aGUgY2lyY29zIHBsb3QuIn0KCmxpc3QoaWQxX2F0YWNfcGVha3MsIGlkMV9nZW5lKSAlPiUgCiAgICBkb3dubG9hZHRoaXM6OmRvd25sb2FkX3RoaXMoCiAgICBvdXRwdXRfbmFtZSA9ICJGaWd1cmUzQSBkYXRhIiwKICAgIG91dHB1dF9leHRlbnNpb24gPSAiLnhsc3giLAogICAgYnV0dG9uX2xhYmVsID0gIkRvd25sb2FkIEZpZ3VyZSAzQSBkYXRhIGFzIHhsc3giLAogICAgYnV0dG9uX3R5cGUgPSAicHJpbWFyeSIsCiAgICBoYXNfaWNvbiA9IFRSVUUsCiAgICBpY29uID0gImZhIGZhLXNhdmUiCiAgKQoKYGBgCgo8YnI+CgojIyMjIEZpZ3VyZSBTM0E6IENvcnJlbGF0aW9uIGJldHdlZW4gcHJvdGVpbiBhYnVuZGFuY2UgYW5kIGNocm9tYXRpbiBhY2Nlc3NpYmlsaXR5IGFjcm9zcyB0aGUgZ2Vub21lCgpgYGB7ciBGaWd1cmVTM0EsIGZpZy5jYXA9IkZpZ3VyZSAzQTogQSBoZWF0bWFwIG9mIFBlYXJzb24gY29ycmVsYXRpb24gY29lZmZpY2llbnRzIGJldHdlZW4gcHJvdGVpbiBhYnVuZGFuY2UgYW5kIGNocm9tYXRpbiBhY2Nlc3NpYmlsaXR5IGFjcm9zcyB0aGUgZ2Vub21lLiBQcm90ZWlucyBlbmNvZGVkIG9uIHRoZSBzZXggY2hyb21vc29tZXMgd2VyZSBleGNsdWRlZCBmcm9tIHRoZSBhbmFseXNpcyB0byBsaW1pdCBzZXggZWZmZWN0cyBkdWUgdG8gWCBnZW5lIGRvc2FnZS4gQ29ycmVsYXRpb24gYmV0d2VlbiBhbGwgYXV0b3NvbWFsIHByb3RlaW5zIGFuZCBhY2Nlc3NpYmlsaXR5IGF0IEFUQUMtc2VxIHBlYWtzIHdlcmUgY2FsY3VsYXRlZC4gRm9yIHBsb3R0aW5nLCBwcm90ZWlucyBhbmQgY2hyb21hdGluIHJlZ2lvbnMgYXJlIGdyb3VwZWQgaW4gNSBLYiBiaW5zIGFuZCB0aGUgcG9pbnRzIGFyZSBjb2xvcmVkIGFuZCBzaXplZCBieSB0aGUgbWF4aW11bSBjb3JyZWxhdGlvbiB2YWx1ZSBpbiBlYWNoIGJpbi4ifQoKa25pdHI6OmluY2x1ZGVfZ3JhcGhpY3MoIGhlcmUoIm1lYW5fc2hhcmVkUHJvdF9tYXhBVEFDXzA1TWJfc2NhbGVkLnBuZyIpKQoKYGBgCgo8YnI+CgojIyMjIExpc3Qgb2YgY2hyb21hdGluIHJlZ2lvbnMgc2hvd2luZyBoaWdoIGNvcnJlbGF0aW9uIHRvIHByb3RlaW4gYWJ1bmRhbmNlIGFjcm9zcyB0aGUgZ2Vub21lCgpgYGB7ciwgZmlnLmNhcD0iRnVsbCBsaXN0IG9mIGNocm9tYXRpbiByZWdpb25zIHRoYXQgc2hvdyBoaWdoIGNvcnJlbGF0aW9uICggYWJzKGNvcikgPjAuNSkgdG8gcHJvdGVpbnMgYWNyb3NzIHRoZSBnZW5vbWUgd2l0aCBhbm5vdGF0aW9ucy4ifQoKcHJvdC5jb3IuYXRhY3BlYWtzICU+JQogIGZpbHRlcihuID4gMTAwKSAlPiUKICAjZmlsdGVyKCFjb3JfZ2VuZSAlaW4lIChmaWx0ZXIocm5hLmNvci5nZW5lcywgbiA+IDUwKSkkbWdpX3N5bWJvbCkgJT4lCiAgc2VsZWN0KGNvcl9nZW5lLCBwZWFrX2lkLCBjb3JyLCBhbm5vdGF0aW9uLCBtZ2lfc3ltYm9sLCBnZW5lX3N0YXJ0LCBnZW5lX2VuZCwgZ2VuZV9jaHIsIG4pICU+JQogIHJlbmFtZShgIyBvZiBjb3JyZWFsYXRlZCBBVEFDLXNlcSBwZWFrc2AgPSBuKSAlPiUKICBtdXRhdGUoY29yciA9IGZvcm1hdEMoY29yciwgZm9ybWF0ID0gImciLCBkaWdpdHMgPSAyKSkgJT4lCiAgZ3JvdXBfYnkocGVha19pZCkgJT4lCiAgbXV0YXRlKG5fZ2VuZSA9IG4oKSkgJT4lCiAgc2VsZWN0KGNvcl9nZW5lLCBwZWFrX2lkLCBjb3JyLCBhbm5vdGF0aW9uLCBtZ2lfc3ltYm9sLCBuX2dlbmUsIGAjIG9mIGNvcnJlYWxhdGVkIEFUQUMtc2VxIHBlYWtzYCkgJT4lCiAgdW5ncm91cCgpICU+JQogIGdyb3VwX2J5KGNvcl9nZW5lKSAlPiUKICB1bmdyb3VwKCkgJT4lCiAgZGlzdGluY3QoKSAlPiUKICBtdXRhdGUoIGNvcl9nZW5lID0gdG91cHBlcihjb3JfZ2VuZSkpICU+JSAKICBzZWxlY3QoIGBQcm90ZWluYD0gKGNvcl9nZW5lKSwKICAgICAgICAgIGBQZWFrIGlkYCA9IHBlYWtfaWQsCiAgICAgICAgICBgQ29ycmVsYXRpb25gID0gY29yciwKICAgICAgICAgIGBQZWFrIGFubm90YXRpb24gKGZ1bmN0aW9uKWAgPSBhbm5vdGF0aW9uLCAKICAgICAgICAgIGBQZWFrIGFubm90YXRpb24gKGdlbmUpYCA9IG1naV9zeW1ib2wsCiAgICAgICAgICBgIyBvZiBjb3JyZWFsYXRlZCBBVEFDLXNlcSBwZWFrc2AsCiAgICAgICAgICBgIyBvZiBjb3JyZWFsYXRlZCBwcm90ZWluc2AgPSBuX2dlbmUKICAgICAgICAgKSAlPiUgCiAgY3JlYXRlX2R0KCkKCgpgYGAKCjxicj4KCiMjIyMgT3Zlci1yZXByZXNlbnRhdGlvbiBvZiB0cmFuc2NyaXB0aW9uIGJpbmRpbmcgc2l0ZXMgaW4gY2hyb21hdGluIHJlZ2lvbnMgd2l0aCBoaWdoIGNvcnJlbGF0aW9uIHRvIHByb3RlaW4gYWJ1bmRhbmNlIGFjcm9zcyB0aGUgZ2Vub21lCgpgYGB7ciBPUkFfQVRBQ19zZXFfcGVha3MsIG1lc3NhZ2U9RkFMU0UsIHdhcm5pbmc9RkFMU0UsIGZpZy5jYXA9Ik92ZXItcmVwcmVzZW50ZWQgdHJhbnNjcmlwdGlvbiBmYWN0b3IgYmluZGluZyBzaXRlcyBpbiBjaHJvbWF0aW4gcmVnaW9ucyBzaG93aW5nIGhpZ2ggY29ycmVsYXRpb24gdG8gYWJ1bmRhbmNlIG9mIHByb3RlaW5zIHNlZW4gYXMgaG9yaXpvbnRhbCBiYW5kcyBpbiBGaWd1cmUgUzNBLiJ9CgpiYWNrZ3JvdW5kX2F0YWNfcGVha3MgPC0gIHRpYmJsZSggcGVha19pZCA9IGF0YWMucGVhay5hbm5vdHNfZnVsbCRwZWFrX2lkICkgJT4lCiAgc2VwYXJhdGUoIHBlYWtfaWQsIGludG8gPSBjKCJDaHIiLCAiU3RhcnQiLCJFbmQiKSwgcmVtb3ZlID0gRkFMU0UpICU+JQogIG11dGF0ZSggQ2hyID0gZ3N1YigicGVhayIsImNociIsQ2hyKSkgJT4lIAogIG1ha2VHUmFuZ2VzRnJvbURhdGFGcmFtZSguLAogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAga2VlcC5leHRyYS5jb2x1bW5zID0gRiwKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHNlcW5hbWVzLmZpZWxkID0gYygiQ2hyIiksCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBzdGFydC5maWVsZCA9ICJTdGFydCIsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBlbmQuZmllbGQgPSAiRW5kIikKCnByb3QuY29yLmF0YWNwZWFrcyAlPiUKICBmaWx0ZXIobiA+IDEwMCkgJT4lCiAgdW5ncm91cCgpICU+JQogIGZpbHRlciggY29yciA+IDApICU+JSAKICBsZWZ0X2pvaW4oLiwgc2VsZWN0KGFsbC5wcm90cywgImNvcl9nZW5lX2lkIiA9ICJwcm90ZWluX2lkIiwgImNvcl9nZW5lIiA9ICJtZ2lfc3ltYm9sIikpICU+JQogIGxlZnRfam9pbihhdGFjLnBlYWsuYW5ub3RzKSAlPiUKICBzZWxlY3QoY29yX2dlbmVfaWQsIGNvcl9nZW5lLCBwZWFrX2lkLCBjb3JyLCBhbm5vdGF0aW9uLCBtZ2lfc3ltYm9sKSAlPiUKICBtdXRhdGUoY29yciA9IGZvcm1hdEMoY29yciwgZm9ybWF0ID0gImciLCBkaWdpdHMgPSAyKSkgJT4lCiAgYXJyYW5nZShjb3JfZ2VuZSkgJT4lCiAgZmlsdGVyKCFpcy5uYShtZ2lfc3ltYm9sKSkgJT4lCiAgYXNfdGliYmxlKCkgJT4lCiAgc2VsZWN0KGNvcl9nZW5lLCBwZWFrX2lkKSAlPiUKICAjZmlsdGVyKGNvcl9nZW5lID09IkFoZGMxIikgJT4lCiAgc2VwYXJhdGUoIHBlYWtfaWQsIGludG8gPSBjKCJDaHIiLCAiU3RhcnQiLCJFbmQiKSwgcmVtb3ZlID0gRkFMU0UpICU+JQogIG11dGF0ZSggQ2hyID0gZ3N1YigicGVhayIsImNociIsQ2hyKSkgJT4lCiAgZ3JvdXBfYnkoY29yX2dlbmUpICU+JQogIG5lc3QoKSAlPiUKICBtdXRhdGUoZ2VuZVNldCA9IG1hcChkYXRhLCBmdW5jdGlvbihkZikgewogICAgZ2VuZXMgPSBtYWtlR1Jhbmdlc0Zyb21EYXRhRnJhbWUoIGRhdGEsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBrZWVwLmV4dHJhLmNvbHVtbnMgPSBGLAogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgc2VxbmFtZXMuZmllbGQgPSBjKCJDaHIiKSwKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHN0YXJ0LmZpZWxkID0gIlN0YXJ0IiwKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGVuZC5maWVsZCA9ICJFbmQiKQogICAgcmV0dXJuKGdlbmVzKQoKICB9KSkgJT4lCiAgc2VsZWN0KC1kYXRhKSAlPiUKICB1bmdyb3VwKCkgJT4lCiAgbXV0YXRlKCB1c2VyU2V0ID0gc2VxKDE6bigpKSwgdHlwZSA9InBvcyIpLT4gYWxsX3Byb3RfYXRhY19wZWFrc19wb3MKCnByb3QuY29yLmF0YWNwZWFrcyAlPiUKICBmaWx0ZXIobiA+IDEwMCkgJT4lCiAgdW5ncm91cCgpICU+JQogIGZpbHRlciggY29yciA8IDApICU+JSAKICAjZmlsdGVyKCFjb3JfZ2VuZSAlaW4lIChmaWx0ZXIocm5hLmNvci5nZW5lcywgbiA+IDUwKSkkbWdpX3N5bWJvbCkgJT4lCiAgbGVmdF9qb2luKC4sIHNlbGVjdChhbGwucHJvdHMsICJjb3JfZ2VuZV9pZCIgPSAicHJvdGVpbl9pZCIsICJjb3JfZ2VuZSIgPSAibWdpX3N5bWJvbCIpKSAlPiUKICBsZWZ0X2pvaW4oYXRhYy5wZWFrLmFubm90cykgJT4lCiAgc2VsZWN0KGNvcl9nZW5lX2lkLCBjb3JfZ2VuZSwgcGVha19pZCwgY29yciwgYW5ub3RhdGlvbiwgbWdpX3N5bWJvbCkgJT4lCiAgbXV0YXRlKGNvcnIgPSBmb3JtYXRDKGNvcnIsIGZvcm1hdCA9ICJnIiwgZGlnaXRzID0gMikpICU+JQogIGFycmFuZ2UoY29yX2dlbmUpICU+JQogIGZpbHRlcighaXMubmEobWdpX3N5bWJvbCkpICU+JQogIGFzX3RpYmJsZSgpICU+JQogIHNlbGVjdChjb3JfZ2VuZSwgcGVha19pZCkgJT4lCiAgI2ZpbHRlcihjb3JfZ2VuZSA9PSJBaGRjMSIpICU+JQogIHNlcGFyYXRlKCBwZWFrX2lkLCBpbnRvID0gYygiQ2hyIiwgIlN0YXJ0IiwiRW5kIiksIHJlbW92ZSA9IEZBTFNFKSAlPiUKICBtdXRhdGUoIENociA9IGdzdWIoInBlYWsiLCJjaHIiLENocikpICU+JQogIGdyb3VwX2J5KGNvcl9nZW5lKSAlPiUKICBuZXN0KCkgJT4lCiAgbXV0YXRlKGdlbmVTZXQgPSBtYXAoZGF0YSwgZnVuY3Rpb24oZGYpIHsKICAgIGdlbmVzID0gbWFrZUdSYW5nZXNGcm9tRGF0YUZyYW1lKCBkYXRhLAogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAga2VlcC5leHRyYS5jb2x1bW5zID0gRiwKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHNlcW5hbWVzLmZpZWxkID0gYygiQ2hyIiksCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBzdGFydC5maWVsZCA9ICJTdGFydCIsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBlbmQuZmllbGQgPSAiRW5kIikKICAgIHJldHVybihnZW5lcykKCiAgfSkpICU+JQogIHNlbGVjdCgtZGF0YSkgJT4lCiAgdW5ncm91cCgpICU+JQogIG11dGF0ZSggdXNlclNldCA9IHNlcSgxOm4oKSksIHR5cGUgPSJuZWciKS0+IGFsbF9wcm90X2F0YWNfcGVha3NfbmVnCgpnZW5lc1NldHNfcG9zIDwtIEdSYW5nZXNMaXN0KGMoYWxsX3Byb3RfYXRhY19wZWFrc19wb3MkZ2VuZVNldCkKKQpnZW5lc1NldHNfbmVnIDwtIEdSYW5nZXNMaXN0KGMoYWxsX3Byb3RfYXRhY19wZWFrc19uZWckZ2VuZVNldCkKKQoKb3JhX3Byb3RfdW5pcXVlX2F0YWNfcGVha3NfcG9zIDwtIHJ1bkxPTEEoZ2VuZXNTZXRzX3BvcywKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGJhY2tncm91bmRfYXRhY19wZWFrcywKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJlZ2lvbkRCLAogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgY29yZXM9MSkKCm9yYV9wcm90X3VuaXF1ZV9hdGFjX3BlYWtzX25lZyA8LSBydW5MT0xBKGdlbmVzU2V0c19uZWcsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBiYWNrZ3JvdW5kX2F0YWNfcGVha3MsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICByZWdpb25EQiwKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvcmVzPTEpCgpvcmFfcHJvdF91bmlxdWVfYXRhY19wZWFrc19wb3MkcVZhbHVlIDwtIChxdmFsdWUoIDEwXigtb3JhX3Byb3RfdW5pcXVlX2F0YWNfcGVha3NfcG9zJHBWYWx1ZUxvZyApKSkkcXZhbHVlcwoKb3JhX3Byb3RfdW5pcXVlX2F0YWNfcGVha3NfbmVnJHFWYWx1ZSA8LSAocXZhbHVlKCAxMF4oLW9yYV9wcm90X3VuaXF1ZV9hdGFjX3BlYWtzX25lZyRwVmFsdWVMb2cgKSkpJHF2YWx1ZXMKCgpvcmFfcHJvdF91bmlxdWVfYXRhY19wZWFrc19uZWcgJT4lIAogIGxlZnRfam9pbiggc2VsZWN0KGFsbF9wcm90X2F0YWNfcGVha3NfbmVnLCB1c2VyU2V0LCBjb3JfZ2VuZSkgKSAlPiUgCiAgZmlsdGVyKCBxVmFsdWUgPCAwLjA1KSAlPiUKICBmaWx0ZXIoIGNlbGxUeXBlICVpbiUgYygiRW1icnlvbmljIFN0ZW0gQ2VsbCIsICJFUy1CcnVjZTQiLCJFbWJ5b25pYyBzdGVtIGNlbGwiLCJFbWJyeW9uaWMgU3RlbSBDZWxscyIsICJFbWJyeW9uaWMgc3RlbSBjZWxscyIsICJlbWJyeW9uaWMgc3RlbSBjZWxscyIsImVtYnJ5b25pYyBzdGVtIGNlbGwiKSApICU+JSAKICBncm91cF9ieShjb3JfZ2VuZSApICU+JSAKICBtdXRhdGUoIEdlbmVzID0gcGFzdGUoIHVuaXF1ZShhbnRpYm9keSksIGNvbGxhcHNlID0gIiwgIiksCiAgICAgICAgICBuID0gbl9kaXN0aW5jdChhbnRpYm9keSkpICU+JQogIGFycmFuZ2UoIGRlc2MobikpICU+JSAKICBtdXRhdGUoIGNvcl9nZW5lID0gdG91cHBlcihjb3JfZ2VuZSkpICU+JSAKICBzZWxlY3QoIGBQcm90ZWluYD0gKGNvcl9nZW5lKSwKICAgICAgICAgIGBPdmVycmVwcmVzZW50ZWQgVEYgYmluZGluZyBzaXRlcyBpbiBuZWdhdGl2ZWx5IGNvcnJlbGF0ZWQgQVRBQy1zZXEgcGVha3NgID0gR2VuZXMpICU+JQogIGRpc3RpbmN0KCkgJT4lIAogIGZ1bGxfam9pbigKICAgIG9yYV9wcm90X3VuaXF1ZV9hdGFjX3BlYWtzX3BvcyAlPiUgCiAgICAgIGxlZnRfam9pbiggc2VsZWN0KGFsbF9wcm90X2F0YWNfcGVha3NfcG9zLCB1c2VyU2V0LCBjb3JfZ2VuZSkgKSAlPiUgCiAgICAgIGZpbHRlciggcVZhbHVlIDwgMC4wNSkgJT4lCiAgICAgIGZpbHRlciggY2VsbFR5cGUgJWluJSBjKCJFbWJyeW9uaWMgU3RlbSBDZWxsIiwgIkVTLUJydWNlNCIsIkVtYnlvbmljIHN0ZW0gY2VsbCIsIkVtYnJ5b25pYyBTdGVtIENlbGxzIiwgIkVtYnJ5b25pYyBzdGVtIGNlbGxzIiwgImVtYnJ5b25pYyBzdGVtIGNlbGxzIiwiZW1icnlvbmljIHN0ZW0gY2VsbCIpICkgJT4lIAogICAgICBncm91cF9ieSggY29yX2dlbmUpICU+JSAKICAgICAgbXV0YXRlKCBHZW5lcyA9IHBhc3RlKCB1bmlxdWUoYW50aWJvZHkpLCBjb2xsYXBzZSA9ICIsICIpLAogICAgICAgICAgbiA9IG5fZGlzdGluY3QoYW50aWJvZHkpKSAlPiUKICAgICAgYXJyYW5nZSggZGVzYyhuKSkgJT4lIAogICAgICBtdXRhdGUoY29yX2dlbmUgPSB0b3VwcGVyKGNvcl9nZW5lKSkgJT4lIAogICAgICBzZWxlY3QoIGBQcm90ZWluYD0gKGNvcl9nZW5lKSwKICAgICAgICAgIGBPdmVycmVwcmVzZW50ZWQgVEYgYmluZGluZyBzaXRlcyBpbiBwb3NpdGl2ZWx5IGNvcnJlbGF0ZWQgQVRBQy1zZXEgcGVha3NgID0gR2VuZXMpICU+JQogICAgICBkaXN0aW5jdCgpCiAgKSAtPiBhbGxfbG9sYV9yZXN1bHRzCgphbGxfbG9sYV9yZXN1bHRzICU+JSAKICBjcmVhdGVfZHQoKQoKYGBgCgo8YnI+CgojIyMjIFRhYmxlIFM0OiBMaXN0IG9mIHByb3RlaW5zIHdpdGggaGlnaCBjb3JyZWxhdGlvbiB0byBBVEFDLXNlcSBwZWFrcyBnZW5vbWUgd2lkZS4gCgpUaGUgdGFibGUgaW5jbHVkZXMgYWRkaXRpb25hbCBhbm5vdGF0aW9ucyBzdWNoIGFzIGNlbGx1bGFyIGxvY2F0aW9uLCBJbnRlcnBybyBkb21haW5zLCBvdmVyLXJlcHJlc2VudGVkIHRyYW5zY3JpcHRpb24gZmFjdG9yIGJpbmRpbmcgc2l0ZXMgaW4gQVRBQy1zZXEgcGVha3Mgd2l0aCBuZWdhdGl2ZSBhbmQgcG9zaXRpdmUgY29ycmVsYXRpb25zLCBhbmQgcmVsZXZhbnQgcmVmZXJlbmNlcyBoaWdobGlnaHRpbmcgcm9sZXMgaW4gcGx1cmlwb3RlbmN5IHJlZ3VsYXRpb24gZm9yIGVhY2ggcHJvdGVpbi4KCmBgYHtyIFRhYmxlUzQsIGV2YWwgPSBGQUxTRX0KCiMgZ2V0IHRoZSBsaXN0IG9mIHByb3RlaW5zIHRoYXQgc2hvdyBoaWdoIGNvcnJlbGF0aW9uIChhYnMoY29yKT4wLjUpIHRvIGF0IGxlYXN0IDEwMCBBVEFDLXNlcSBwZWFrcwphbGxfYW5ub3Rfdjk4X3dHTyA8LSByZWFkX3RzdiggaGVyZSgiLi4vcFFUTF93ZWJzaXRlL19kYXRhIiwiZW5zZW1ibF9nZW5lX2Fubm90YXRpb25zX3Y5OF93R08udHh0IikpICU+JSAKcmVuYW1lKCAiZW5zZW1ibF9nZW5lX2lkIiA9ICJHZW5lIHN0YWJsZSBJRCIsCiAgICAgICAgICAicHJvdGVpbl9pZCIgPSAiUHJvdGVpbiBzdGFibGUgSUQiLAogICAgICAgICAgImdlbmVfc3RhcnQiID0gIkdlbmUgc3RhcnQgKGJwKSIsCiAgICAgICAgICAiZ2VuZV9lbmQiID0gIkdlbmUgZW5kIChicCkiLAogICAgICAgICAgImdlbmVfY2hyIiA9ICJDaHJvbW9zb21lL3NjYWZmb2xkIG5hbWUiLAogICAgICAgICAgIm1naV9zeW1ib2wiID0gIk1HSSBzeW1ib2wiLAogICAgICAgICAgImdlbmVfYmlvdHlwZSIgPSAiR2VuZSB0eXBlIiwKICAgICAgICAiR09fdGVybV9uYW1lIj0iR08gdGVybSBuYW1lIiwKICAgICAgICAiR09fdGVybV9kZWYiID0gIkdPIHRlcm0gZGVmaW5pdGlvbiIsCiAgICAgICAgIkdPX2RvbWFpbiIgPSAiR08gZG9tYWluIiwKICAgICAgICAiR09fSUQiPSAiR08gdGVybSBhY2Nlc3Npb24iCiAgICAgICAgKQoKYWxsX2Fubm90X3c5OF93aW50ZXJwcm8gPC0gcmVhZF90c3YoIGhlcmUoIi4uL3BRVExfd2Vic2l0ZS9fZGF0YSIsImVuc2VtYmxfdjk4X2ludGVycHJvLnR4dCIpKSAlPiUgCiAgcmVuYW1lKCAiZW5zZW1ibF9nZW5lX2lkIiA9ICJHZW5lIHN0YWJsZSBJRCIsCiAgICAgICAgICAicHJvdGVpbl9pZCIgPSAiUHJvdGVpbiBzdGFibGUgSUQiKSAlPiUgCiAgc2VsZWN0KHByb3RlaW5faWQsIGludGVycHJvX2RvbWFpbnMgPWBJbnRlcnBybyBEZXNjcmlwdGlvbmAgKSAlPiUgCiAgZGlzdGluY3QoKQogIAoKcHJvdC5jb3IuYXRhY3BlYWtzICU+JSAKICBmaWx0ZXIobiA+IDEwMCkgJT4lCiAgdW5ncm91cCgpICU+JQogIG11dGF0ZSggY29yX3R5cGUgPSBpZmVsc2UoIGNvcnIgPiAwLCAiUG9zaXRpdmUiLCAiTmVnYXRpdmUiKSkgJT4lIAogIGdyb3VwX2J5KCBjb3JfZ2VuZSwgY29yX3R5cGUpICU+JSAKICBjb3VudCgpICU+JSAKICBwaXZvdF93aWRlciggY29yX2dlbmUsIG5hbWVzX2Zyb20gPSAiY29yX3R5cGUiLCB2YWx1ZXNfZnJvbSA9ICJuIikgJT4lIAogIG11dGF0ZSggc3VtID0gUG9zaXRpdmUgKyBOZWdhdGl2ZSkgJT4lIAogIG11dGF0ZSggYE51bWJlciBvZiBjb3JyZWxhdGVkIEFUQUMtc2VxIHBlYWtzIChwb3NpdGl2ZSAvIG5lZ2F0aXZlKWAgPSBzdHJfYyhzdW0sICIgKCIsUG9zaXRpdmUsIi8iLCBOZWdhdGl2ZSwiKSAiKSkgJT4lIAogIHNlbGVjdCggY29yX2dlbmUsYE51bWJlciBvZiBjb3JyZWxhdGVkIEFUQUMtc2VxIHBlYWtzIChwb3NpdGl2ZSAvIG5lZ2F0aXZlKWApICAtPiBwZWFrX251bXMKCnByb3QuY29yLmdlbmVzICU+JSAKICBmaWx0ZXIobiA+IDEwMCkgJT4lIAogICMgYWRkIGNlbGx1bGFyIGxvY2F0aW9uIAogIGxlZnRfam9pbiggICBhbGxfYW5ub3Rfdjk4X3dHTyAlPiUKICAgICAgICAgICAgICAgZmlsdGVyKCBHT19kb21haW4gPT0iY2VsbHVsYXJfY29tcG9uZW50IikgJT4lIAogICAgICAgICAgICAgICBzZWxlY3QoIHByb3RlaW5faWQsIAogICAgICAgICAgICAgICAgICAgICAgIEdPX3Rlcm1fbmFtZQogICAgICAgICAgICAgICAgICAgICAgICkKICAgICAgICAgICAgICkgJT4lIAogIGdyb3VwX2J5KG1naV9zeW1ib2wsIHByb3RlaW5faWQsIGdlbmVfY2hyLCBuKSAlPiUgCiAgc3VtbWFyaXNlKGFjcm9zcyhHT190ZXJtX25hbWUsIHN0cl9jLCBjb2xsYXBzZT0iIDsgIikpIC0+IHByb3RfbG9jYXRpb25zIAogIApwcm90LmNvci5nZW5lcyAlPiUgCiAgZmlsdGVyKG4gPiAxMDApICU+JSAKICAjIGFkZCBpbnRlcnBybyBkb21haW5zIAogIGxlZnRfam9pbiggYWxsX2Fubm90X3c5OF93aW50ZXJwcm8pICU+JSAKICBncm91cF9ieShtZ2lfc3ltYm9sLCBwcm90ZWluX2lkLCBnZW5lX2NociwgbikgJT4lIAogIHN1bW1hcmlzZShhY3Jvc3MoaW50ZXJwcm9fZG9tYWlucywgc3RyX2MsIGNvbGxhcHNlPSIgOyAiKSkgLT4gcHJvdF9pbnRlcnByb19kb21zCgpwZWFrX251bXMgJT4lIAogIHJlbmFtZShtZ2lfc3ltYm9sID0gY29yX2dlbmUpICU+JSAKICBsZWZ0X2pvaW4oIHByb3RfbG9jYXRpb25zKSAlPiUgCiAgbGVmdF9qb2luKCBwcm90X2ludGVycHJvX2RvbXMpICU+JSAKICBtdXRhdGUobWdpX3N5bWJvbCA9IHRvdXBwZXIobWdpX3N5bWJvbCksCiAgICAgICAgIGdlbmVfY2hyID0gYXMubnVtZXJpYyhnZW5lX2NocikpICU+JSAKICBzZWxlY3QoYFByb3RlaW4gSURgID0gcHJvdGVpbl9pZCwgCiAgICAgICAgIGBQcm90ZWluYCA9IChtZ2lfc3ltYm9sKSwgCiAgICAgICAgIGBQcm90ZWluIGxvY2F0aW9uIChDaHIpYCA9IGdlbmVfY2hyLCAKICAgICAgICAgYENlbGx1bGFyIGxvY2F0aW9uYCA9IEdPX3Rlcm1fbmFtZSwKICAgICAgICAgYEludGVycHJvIGRvbWFpbmAgPSBpbnRlcnByb19kb21haW5zLAogICAgICAgICBgTnVtYmVyIG9mIGNvcnJlbGF0ZWQgQVRBQy1zZXEgcGVha3MgKHBvc2l0aXZlIC8gbmVnYXRpdmUpYAogICAgICAgICApICU+JQogIGxlZnRfam9pbigKICAgIGFsbF9sb2xhX3Jlc3VsdHMKICApICU+JSAKICAjIEFoZGMxIGFubm90YXRpb24gaXMgbWlzc2luZywgZml4aW5nIHRoYXQKICBtdXRhdGUoIGBDZWxsdWxhciBsb2NhdGlvbmAgPSBpZmVsc2UoIFByb3RlaW4gPT0iQUhEQzEiLCAibnVjbGV1cyIsIGBDZWxsdWxhciBsb2NhdGlvbmApKSAlPiUgCiAgIyBhZGQgb2JzZXJ2ZWQgYXQgdGhlIHRyYW5zY3JpcHQgbGV2ZWw/CiAgbXV0YXRlKGBPYnNlcnZlZCBhdCB0aGUgdHJhbnNjdHJpcHQgbGV2ZWw/YCA9IGlmZWxzZSggCiAgICBQcm90ZWluICVpbiUgYygiQVJIR0VGMSIsIkdKQjMiLCJOQVBSVCIsICJPT0VQIiwgIlBITERBMiIsICJQVVM3TCIpLAogICAgIlllcyIsICJObyIpKSAlPiUgCiAgIyBhZGQgcmVmZXJlbmNlcwogIG11dGF0ZSggCiAgICBgUHVibGlzaGVkIHJvbGUgaW4gcGx1cmlwb3RlbmN5IFtyZWZdYCA9IGNhc2Vfd2hlbigKICAgICAgUHJvdGVpbiA9PSAiQUhEQzEifiJNb3JlaXJhIFMsIFNlbyBDLCBHb3Jkb24gViwgWGluZyBTLCBXdSBSLCBQb2xlbmEgRSwgZXQgYWwuIEVuZG9nZW5vdXMgQmlvSUQgZWx1Y2lkYXRlcyBUQ0Y3TDEgaW50ZXJhY3RvbWUgbW9kdWxhdGlvbiB1cG9uIEdTSy0zIGluaGliaXRpb24gaW4gbW91c2UgRVNDcy4gMjAxOCBPY3QgcC4gNDMxMDIzLiBkb2k6MTAuMTEwMS80MzEwMjMiLAogICAgICBQcm90ZWluID09IklEMSJ+IlJvbWVyby1MYW5tYW4sIEUuRS4sIFBhdmxvdmljLCBTLiwgQW1sYW5pLCBCLiwgQ2hpbiwgWS4sIGFuZCBCZW5lenJhLCBSLiAoMjAxMikuIElkMSBNYWludGFpbnMgRW1icnlvbmljIFN0ZW0gQ2VsbCBTZWxmLVJlbmV3YWwgYnkgVXAtUmVndWxhdGlvbiBvZiBOYW5vZyBhbmQgUmVwcmVzc2lvbiBvZiBCcmFjaHl1cnkgRXhwcmVzc2lvbi4gU3RlbSBDZWxscyBEZXYuIDIxLCAzODTigJMzOTMuIiwKICAgICAgUHJvdGVpbiA9PSJVSFJGMiJ+IldhbGtlciBFLCBDaGFuZyBXWSwgSHVua2FwaWxsZXIgSiwgQ2FnbmV5IEcsIEdhcmNoYSBLLCBUb3JjaGlhIEosIEtyb2dhbiBOSiwgUmVpdGVyIEpGLCBTdGFuZm9yZCBXTC4gUG9seWNvbWItbGlrZSAyIGFzc29jaWF0ZXMgd2l0aCBQUkMyIGFuZCByZWd1bGF0ZXMgdHJhbnNjcmlwdGlvbmFsIG5ldHdvcmtzIGR1cmluZyBtb3VzZSBlbWJyeW9uaWMgc3RlbSBjZWxsIHNlbGYtcmVuZXdhbCBhbmQgZGlmZmVyZW50aWF0aW9uLiBDZWxsIFN0ZW0gQ2VsbC4gMjAxMCBGZWIgNTs2KDIpOjE1My02Ni4gZG9pOiAxMC4xMDE2L2ouc3RlbS4yMDA5LjEyLjAxNC4gUE1JRDogMjAxNDQ3ODg7IFBNQ0lEOiBQTUMyODQ5MDA0LiIKICAgICkpIC0+IHRhYmxlX3M0Cgp3cml0ZXhsOjp3cml0ZV94bHN4KCB0YWJsZV9zNCwKICAgICAgICAgICAgICAgICAgICBwYXRoID0gaGVyZSgiVGFibGVTNF9Qcm90ZWluc193X2NvcnJfdG9BVEFDc2VxLnhsc3giKSwKICAgICAgICAgICAgICAgICAgICBjb2xfbmFtZXMgPSBUUlVFLAogICAgICAgICAgICAgICAgICAgIGZvcm1hdF9oZWFkZXJzID0gVFJVRQogICAgICAgICAgICAgICAgICAgICkKCmBgYAoKYGBge3IgVGFibGVTNF9kaXNwbGF5LCAgZWNobyA9RkFMU0V9CgojeGZ1bjo6ZW1iZWRfZmlsZShoZXJlKCJUYWJsZV9TNC54bHN4IikpCgpkb3dubG9hZF9maWxlKAogIHBhdGggPSBoZXJlKCJUYWJsZV9TNC54bHN4IiksCiAgb3V0cHV0X25hbWUgPSAiVGFibGVfUzQiLAogIGJ1dHRvbl9sYWJlbCA9ICJEb3dubG9hZCBUYWJsZV9TNC54bHN4IiwKICBidXR0b25fdHlwZSA9ICJwcmltYXJ5IiwKICBoYXNfaWNvbiA9IFRSVUUsCiAgaWNvbiA9ICJmYSBmYS1zYXZlIiwKICBzZWxmX2NvbnRhaW5lZCA9IEZBTFNFCikKCmBgYAoKPGJyPgo8YnI+CgojIyMgRmlndXJlIDNCOiBDb3ZhcmlhdGlvbiBvZiBwcm90ZW9tZSBhbmQgdHJhbnNjcmlwdG9tZSBhY3Jvc3Mgc2FtcGxlcwoKYGBge3IgRmlndXJlM0JfcHJlcH0KCnNoYXJlZC5wcm90Lm5hbWVzIDwtIHNoYXJlZC5nZW5lcyAlPiUKICBncm91cF9ieShlbnNlbWJsX2dlbmVfaWQpICU+JQogIG11dGF0ZShuZXdfc3ltYm9sID0gcGFzdGUwKG1naV9zeW1ib2wsICJfIiwgMTpuKCkpLAogICAgICAgICBuZXdfZ2VuZV9pZCA9IHBhc3RlMChlbnNlbWJsX2dlbmVfaWQsICJfIiwgMTpuKCkpKQoKc2hhcmVkX3Byb3RfbWF0IDwtICB0KGV4cHJaLmVzY19wcm90W3NoYXJlZC5zYW1wbGVzLCBzaGFyZWQucHJvdC5uYW1lcyRwcm90ZWluX2lkXSkKY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0KSA8LSBwYXN0ZTAoY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0KSwiX3Byb3RlaW4iKQpzaGFyZWRfcm5hX21hdCA8LSAgdChleHByWi5lc2Nfcm5hW3NoYXJlZC5zYW1wbGVzLCBzaGFyZWQucHJvdC5uYW1lcyRlbnNlbWJsX2dlbmVfaWRdKQpjb2xuYW1lcyhzaGFyZWRfcm5hX21hdCkgPC0gcGFzdGUwKGNvbG5hbWVzKHNoYXJlZF9ybmFfbWF0KSwiX3JuYSIpCgpwcm90X3JuYV9zYW1wbGVfY29yIDwtIHJjb3JyKCB4ID0gc2hhcmVkX3Byb3RfbWF0LAogICAgICAgICAgICAgICAgICAgICAgICAgeSA9IHNoYXJlZF9ybmFfbWF0LAogICAgICAgICAgICAgICAgICAgICAgICAgdHlwZSA9ICJwZWFyc29uIikKCgpwcm90X3JuYV9zYW1wbGVfY29yX2RmIDwtIGFzX3RpYmJsZSggcHJvdF9ybmFfc2FtcGxlX2NvciRyW2NvbG5hbWVzKHNoYXJlZF9wcm90X21hdCksIGNvbG5hbWVzKHNoYXJlZF9ybmFfbWF0KV0sCiAgICAgICAgICAgICAgICAgICAgICAgICAgICByb3duYW1lcyA9ICJwcm90ZWluX3NhbXBsZSIpICU+JQogIHBpdm90X2xvbmdlciggY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpLCBuYW1lc190byA9ICJybmFfc2FtcGxlIiwgdmFsdWVzX3RvID0gInIiKSAlPiUKICBpbm5lcl9qb2luKCAoYXNfdGliYmxlKCBwcm90X3JuYV9zYW1wbGVfY29yJFBbY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0KSwgY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpXSwKICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJvd25hbWVzID0gInByb3RlaW5fc2FtcGxlIikgJT4lCiAgICAgIHBpdm90X2xvbmdlciggY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpLCBuYW1lc190byA9ICJybmFfc2FtcGxlIiwgdmFsdWVzX3RvID0gInBfdmFsIikgKSApCgoKcHJvdF9ybmFfc2FtcGxlX2Nvcl9kZiAlPiUKICBtdXRhdGUoIHNhbXBsZWlkX3Byb3QgPSBnc3ViKCJfcHJvdGVpbiIsIiIscHJvdGVpbl9zYW1wbGUpLAogICAgICAgICAgc2FtcGxlaWRfcm5hID0gZ3N1YigiX3JuYSIsIiIsIHJuYV9zYW1wbGUpKSAlPiUKICBmaWx0ZXIoIHNhbXBsZWlkX3JuYSA9PSBzYW1wbGVpZF9wcm90KSAlPiUgc3VtbWFyaXplKCBtZWRfciA9IG1lZGlhbihyKSkgLT4gbWVkaWFuX2Nvcl9ybmFfcHJvdAoKRmlndXJlM0JfZGF0YSA8LSBwcm90X3JuYV9zYW1wbGVfY29yX2RmICU+JQogIG11dGF0ZSggc2FtcGxlaWRfcHJvdCA9IGdzdWIoIl9wcm90ZWluIiwiIixwcm90ZWluX3NhbXBsZSksCiAgICAgICAgICBzYW1wbGVpZF9ybmEgPSBnc3ViKCJfcm5hIiwiIiwgcm5hX3NhbXBsZSkpICU+JQogIGZpbHRlciggc2FtcGxlaWRfcm5hID09IHNhbXBsZWlkX3Byb3QpICU+JQogIHJlbmFtZSggYFNhbXBsZSBpZGAgPSBzYW1wbGVpZF9wcm90LAogICAgICAgICAgYENvcnJlbGF0aW9uYCA9IHIpCgoKYGBgCgoKYGBge3IgRmlndXJlM0IsIG1lc3NhZ2U9RkFMU0UsIHdhcm5pbmc9RkFMU0UsIGNhY2hlPVRSVUUsZmlnLmNhcCA9ICJGaWd1cmUgM0I6IERPIG1FU0NzIHNob3cgYSB3aWRlIHJhbmdlIG9mIGNvcnJlbGF0aW9ucyBiZXR3ZWVuIHRoZWlyIHRyYW5zY3JpcHRvbWUgYW5kIHByb3Rlb21lLiBIaXN0b2dyYW0gb2YgUGVhcnNvbiBjb3JyZWxhdGlvbiBjb2VmZmljaWVudHMgYmV0d2VlbiB0aGUgdHJhbnNjcmlwdG9tZSBhbmQgcHJvdGVvbWUgb2YgRE8gbUVTQyBjZWxsIGxpbmVzIHdpdGggbWF0Y2hpbmcgZ2Vub3R5cGVzIChuID0gMTc0KS4iLCBmaWcuaGVpZ2h0PTMsIGZpZy53aWR0aD01fQoKCkZpZ3VyZTNCX2RhdGEgJT4lIAogIGdncGxvdCgpICsKICBhZXMoeCA9IENvcnJlbGF0aW9uKSArCiAgZ2VvbV9oaXN0b2dyYW0oIHNob3cubGVnZW5kID0gRiwgYmlud2lkdGggPSAwLjAxLCBhbHBoYSA9IDAuOCkgKwogIGdlb21fdmxpbmUoIGFlcyh4aW50ZXJjZXB0ID0gbWVkaWFuKENvcnJlbGF0aW9uLG5hLnJtPVQpKSwgY29sb3IgPSAiYmxhY2siLCBsaW5ldHlwZSA9ICJkYXNoZWQiLCBzaXplID0gMSApKwogIGdlb21fdGV4dCggbWFwcGluZz0gYWVzKAogICAgICAgICAgICAgIHggPSBtZWRpYW4oQ29ycmVsYXRpb24sbmEucm09VCktMC4wOCwKICAgICAgICAgICAgICBsYWJlbCA9IHN0cmluZ3I6OnN0cl93cmFwKHBhc3RlMCgiTWVkaWFuID0gIixyb3VuZChtZWRpYW4oQ29ycmVsYXRpb24sbmEucm09VCksMikpLDcpLCAKICAgICAgICAgICAgKSwKICAgICAgICAgICAgeSA9IDEzLAogICAgICAgICAgICBzaXplID0gNgogICAgICAgICAgCiAgKSsKICB4bGFiKCJDZWxsIGxpbmUgY29ycmVsYXRpb24iKSArCiAgeWxhYigiQ291bnQiKSArCiAgdGhlbWVfcHViY2xlYW4oYmFzZV9zaXplID0gMjApKwogIHhsaW0oMCwwLjYpKwogIHlsaW0oMCwxNSkKCmBgYAoKPGJyPgoKYGBge3IgRmlndXJlM0JfZGF0YSwgZmlnLmNhcD0iQ29ycmVsYXRpb24gdmFsdWVzIHVzZWQgdG8gZ2VuZXJhdGUgRmlndXJlIDNCLiJ9CgpGaWd1cmUzQl9kYXRhICU+JSAKICBzZWxlY3QoCiAgICAgICBgU2FtcGxlIGlkYCAsYENvcnJlbGF0aW9uYCAKICApICU+JSAKICBtdXRhdGVfaWYoaXMubnVtZXJpYywgcm91bmQgLDIpICU+JSAKICBjcmVhdGVfZHQoKQoKYGBgCgo8YnI+CgojIyMjIEZpZ3VyZSBTM0ItQzogVmFyaWF0aW9uIGluIHRyYW5zY3JpcHQgYW5kIHByb3RlaW4gYWJ1bmRhbmNlCgpgYGB7ciBGaWd1cmVTM0JfQywgZmlnLmNhcCA9ICJGaWd1cmUgUzM6IChCLCBDKSBTY2F0dGVycGxvdHMgc2hvd2luZyBtZWFuIGFuZCBjb2VmZmljaWVudCBvZiB2YXJpYXRpb24gKCUgQ1YpIGZvciB0cmFuc2NyaXB0IGFuZCBwcm90ZWluIGFidW5kYW5jZSBmb3IgZ2VuZXMgd2l0aCBib3RoIG1lYXN1cmVtZW50cyAobiA9IDcsMjQxKS4iLCBmaWcud2lkdGg9MTAsIGZpZy5oZWlnaHQ9NSwgd2FybmluZz1GQUxTRSwgbWVzc2FnZT1GQUxTRX0KCiMgZ2V0IG1lYW4gKyAlY3YgZm9yIHByb3RlaW4gYWJ1bmRhbmNlCnZhcl9wcm90IDwtIGV4cHIuZXNjX3Byb3QgJT4lCiAgYXNfdGliYmxlKC4pICU+JQogIHN1bW1hcmlzZV9hbGwobGlzdCh+IHZhciguLCBuYS5ybSA9IFQpKSkgJT4lCiAgcGl2b3RfbG9uZ2VyKGV2ZXJ5dGhpbmcoKSwKICAgICAgICAgICAgICAgIG5hbWVzX3RvID0gInByb3RlaW5faWQiLAogICAgICAgICAgICAgICAgdmFsdWVzX3RvID0idmFyLnByb3QiKSAKCm1lYW5fcHJvdCA8LSBleHByLmVzY19wcm90ICU+JQogIGFzX3RpYmJsZSguKSAlPiUKICBzdW1tYXJpc2VfYWxsKGxpc3QofiBtZWFuKC4sIG5hLnJtID0gVCkpKSAlPiUgCiAgcGl2b3RfbG9uZ2VyKCBldmVyeXRoaW5nKCksCiAgICAgICAgICAgICAgICBuYW1lc190byA9ICJwcm90ZWluX2lkIiwKICAgICAgICAgICAgICAgIHZhbHVlc190byA9Im1lYW4ucHJvdCIpIAoKdmFyX3Byb3QgJT4lIAogIGZ1bGxfam9pbiggbWVhbl9wcm90KSAlPiUgCiAgbXV0YXRlKHNkLnByb3QgPSBzcXJ0KHZhci5wcm90KSkgJT4lCiAgbXV0YXRlKGN2LnByb3QgPSAxMDAgKiBzZC5wcm90IC8gKG1lYW4ucHJvdCkpIC0+IGFsbF92YXJfcHJvdAoKIyBnZXQgbWVhbiArICVjdiBmb3IgdHJhbnNjcmlwdCBhYnVuZGFuY2UKdmFyX3JuYSA8LSBleHByLmVzY19ybmEgJT4lCiAgYXNfdGliYmxlKC4pICU+JQogIHN1bW1hcmlzZV9hbGwobGlzdCh+IHZhciguLCBuYS5ybSA9IFQpKSkgJT4lCiAgcGl2b3RfbG9uZ2VyKCBldmVyeXRoaW5nKCksCiAgICAgICAgICAgICAgICBuYW1lc190byA9ICJlbnNlbWJsX2dlbmVfaWQiLAogICAgICAgICAgICAgICAgdmFsdWVzX3RvID0idmFyLnJuYSIpIAoKbWVhbl9ybmEgPC0gZXhwci5lc2Nfcm5hICU+JQogIGFzX3RpYmJsZSguKSAlPiUKICBzdW1tYXJpc2VfYWxsKGxpc3QofiBtZWFuKC4sIG5hLnJtID0gVCkpKSAlPiUgCiAgcGl2b3RfbG9uZ2VyKCBldmVyeXRoaW5nKCksCiAgICAgICAgICAgICAgICBuYW1lc190byA9ICJlbnNlbWJsX2dlbmVfaWQiLAogICAgICAgICAgICAgICAgdmFsdWVzX3RvID0ibWVhbi5ybmEiKSAKCnZhcl9ybmEgJT4lIAogIGZ1bGxfam9pbiggbWVhbl9ybmEpICU+JSAKICBsZWZ0X2pvaW4oIGFsbC5nZW5lcykgJT4lIAogIG11dGF0ZShzZC5ybmEgPSBzcXJ0KHZhci5ybmEpKSAlPiUKICBtdXRhdGUoY3Yucm5hID0gMTAwICogc2Qucm5hIC8gKG1lYW4ucm5hKSkgLT4gYWxsX3Zhcl9ybmEKCiMgam9pbiB0cmFuc2NyaXB0ICsgcHJvdGVpbiB2YXJpYXRpb24KYWxsX3Zhcl9ybmEgJT4lIAogIGxlZnRfam9pbiggc2hhcmVkLmdlbmVzKSAlPiUgCiAgaW5uZXJfam9pbihhbGxfdmFyX3Byb3QgJT4lIAogICAgICAgICAgICAgICBsZWZ0X2pvaW4oc2hhcmVkLmdlbmVzKSkgLT4gYWxsX3ZhcgoKIyBwbG90dGluZyBmaWd1cmVzIFMzYi1jCmFsbF92YXIgJT4lIAogIGdnc2NhdHRlcigKICAgIC4sCiAgICB4ID0gIm1lYW4ucHJvdCIsIHkgPSAibWVhbi5ybmEiLCBzaXplID0gMywgYWxwaGEgPSAwLjYsCiAgYWRkID0gInJlZy5saW5lIiwgIyBBZGQgcmVncmVzc2lvbiBsaW5lCiAgY29uZi5pbnQgPSBUUlVFLCAjIEFkZCBjb25maWRlbmNlIGludGVydmFsCgogIGFkZC5wYXJhbXMgPSBsaXN0KGNvbG9yID0gImJsdWUiLCBmaWxsID0gImxpZ2h0Z3JheSIpLCBzaG93LmxlZ2VuZC50ZXh0ID0gRkFMU0UsCiAgeXNjYWxlID0gImxvZzEwIgopICsKICBzdGF0X2NvcihtZXRob2QgPSAicGVhcnNvbiIsIGxhYmVsLnggPSAxMCwgbGFiZWwueSA9IDYuMSkgKyAjIEFkZCBjb3JyZWxhdGlvbiBjb2VmZmljaWVudAogICMgc3RhdF9jb3IoICBhZXMobGFiZWwgPSBwYXN0ZSguLnJyLmxhYmVsLi4sIC4ucC5sYWJlbC4uLCBzZXAgPSAifmAsYH4iKSksCiAgIyAgICAgICAgICAgIGxhYmVsLnggPSAwLjY1LCBsYWJlbC55ID0gNikgKyMgQWRkIHJlZ3Jlc3Npb24gUjIKICB4bGFiKCJNZWFuIHByb3RlaW4gYWJ1bmRhbmNlIikgKwogIHlsYWIoIk1lYW4gdHJhbnNjcmlwdCBhYnVuZGFuY2UiKSArCiAgdGhlbWVfcHViY2xlYW4oYmFzZV9zaXplID0gMTgpICsgcnJlbW92ZSgibGVnZW5kIikgLT4gZmlndXJlX3MzYgpmaWd1cmVfczNiIDwtIGdncGFyKGZpZ3VyZV9zM2IsIHhsaW0gPSBjKDUsIDE1KSkKCmFsbF92YXIgJT4lIAogIGdnc2NhdHRlciguICwKICAgIHggPSAiY3YucHJvdCIsIHkgPSAiY3Yucm5hIiwgc2l6ZSA9IDMsIGFscGhhID0gMC42LAogICAgYWRkID0gInJlZy5saW5lIiwgIyBBZGQgcmVncmVzc2lvbiBsaW5lCiAgICBjb25mLmludCA9IFRSVUUsICMgQWRkIGNvbmZpZGVuY2UgaW50ZXJ2YWwKICAKICAgIGFkZC5wYXJhbXMgPSBsaXN0KGNvbG9yID0gImJsdWUiLCBmaWxsID0gImxpZ2h0Z3JheSIpLCBzaG93LmxlZ2VuZC50ZXh0ID0gRkFMU0UsCiAgICB5c2NhbGUgPSAibG9nMTAiLCAKICAgIHhzY2FsZSA9ICJsb2cxMCIKICApICsKICAgIHN0YXRfY29yKG1ldGhvZCA9ICJwZWFyc29uIiwgbGFiZWwueCA9IDAuOSwgbGFiZWwueSA9IDMuMDIpICsgIyBBZGQgY29ycmVsYXRpb24gY29lZmZpY2llbnQKICAgICMgc3RhdF9jb3IoICBhZXMobGFiZWwgPSBwYXN0ZSguLnJyLmxhYmVsLi4sIC4ucC5sYWJlbC4uLCBzZXAgPSAifmAsYH4iKSksCiAgICAjICAgICAgICAgICAgbGFiZWwueCA9IC0wLjAxLCBsYWJlbC55ID0gMykgKyMgQWRkIHJlZ3Jlc3Npb24gUjIKICAgIHhsYWIoIiUgQ1YgcHJvdGVpbiBhYnVuZGFuY2UiKSArCiAgICB5bGFiKCIlIENWIHRyYW5zY3JpcHQgYWJ1bmRhbmNlIikgKwogICAgdGhlbWVfcHViY2xlYW4oYmFzZV9zaXplID0gMTgpICsgcnJlbW92ZSgibGVnZW5kIikgLT4gZmlndXJlX3MzYwpmaWd1cmVfczNjIDwtIGdncGFyKCBmaWd1cmVfczNjLCB4bGltPSBjKDEsIDUwKSwgeWxpbSA9IGMoNSwgMTAwMCkpCgpnZ2FycmFuZ2UoZmlndXJlX3MzYiwgCiAgICAgICAgICAgICAgICAgICAgICBmaWd1cmVfczNjLCAKICAgICAgICAgICAgICAgICAgICAgIG5yb3cgPSAxLAogICAgICAgICAgICAgICAgICAgICAgbGFiZWxzID0gYygiQiIsIkMiKSwgCiAgICAgICAgICAgICAgICAgICAgICBmb250LmxhYmVsID0gbGlzdCggc2l6ZSA9IDIwKQogICAgICAgICAgICAgICAgICAgICAgKQoKCmBgYAoKPGJyPgoKIyMjIyBHZW5lcyB3aXRoIGRpZmZlcmVuY2UgaW4gdmFyaWF0aW9uIGluIHRyYW5zY3JpcHQgYW5kIHByb3RlaW4gYWJ1bmRhbmNlCgpgYGB7ciBUYWJsZV9wcm90X3JuYV92YXIsIG1lc3NhZ2U9RkFMU0UsIHdhcm5pbmc9RkFMU0UsIGZpZy5jYXAgPSAiR2VuZXMgd2l0aCBoaWdoIHZhcmlhdGlvbiBpbiB0cmFuc2NyaXB0IGFuZCBsb3cgdmFyaWF0aW9uIGluIHByb3RlaW4gYWJ1bmRhbmNlIGFyZSBvdmVyLXJlcHJlc2VudGVkIGluIHJpYm9zb21hbCBwcm90ZWlucy4iLCBjYWNoZSA9IFRSVUV9CgojIGdlbmVzIHdpdGggaGlnaCB2YXJpYXRpb24gaW4gcHJvdGVpbiBhYnVuZGFuY2UgYW5kIGxvdyB2YXJpYXRpb24gaW4gdHJhbnNjcmlwdCBhYnVuZGFuY2UKYWxsX3ZhciAlPiUKICBmaWx0ZXIoICFpcy5uYShjdi5ybmEpLCAhaXMubmEoY3YucHJvdCkpICU+JQogIGZpbHRlciggY3YucHJvdCA+IHF1YW50aWxlKGN2LnByb3QsIDAuNzUpICYgY3Yucm5hIDwgcXVhbnRpbGUoY3Yucm5hLCAwLjI1KSApICU+JQogIGxlZnRfam9pbihhbGwucHJvdHMpIC0+IHJldl92YXJfcHJvdHMKCiMgZ2VuZXMgd2l0aCBoaWdoIHZhcmlhdGlvbiBpbiB0cmFuc2NyaXB0IGFidW5kYW5jZSBhbmQgbG93IHZhcmlhdGlvbiBpbiBwcm90ZWluIGFidW5kYW5jZQphbGxfdmFyICU+JQogIGZpbHRlciggIWlzLm5hKGN2LnJuYSksICFpcy5uYShjdi5wcm90KSkgJT4lCiAgZmlsdGVyKCBjdi5wcm90IDwgcXVhbnRpbGUoY3YucHJvdCwgMC4yNSkgJiBjdi5ybmEgPiBxdWFudGlsZShjdi5ybmEsIDAuNzUpICkgJT4lCiAgbGVmdF9qb2luKGFsbC5wcm90cykgLT4gcmV2X3Zhcl9wcm90czIKCgpvcmFfcmV2X3Zhcl9wcm90cyA8LSBnb3N0KCBxdWVyeSA9IHVuaXF1ZShyZXZfdmFyX3Byb3RzJG1naV9zeW1ib2wpLAogICAgICAgICAgICAgICAgICAgICBvcmdhbmlzbSA9ICJtbXVzY3VsdXMiLAogICAgICAgICAgICAgICAgICAgICBkb21haW5fc2NvcGUgPSAiY3VzdG9tIiwKICAgICAgICAgICAgICAgICAgICAgY3VzdG9tX2JnID0gc2hhcmVkLmdlbmVzJG1naV9zeW1ib2wsCiAgICAgICAgICAgICAgICAgICAgIGV2Y29kZXMgPSBUUlVFLAogIGNvcnJlY3Rpb25fbWV0aG9kID0gImZkciIKICAgICAgICAgICAgICAgICAgICAgKQpvcmFfcmV2X3Zhcl9wcm90cyRyZXN1bHQgPC0gZmlsdGVyKCBvcmFfcmV2X3Zhcl9wcm90cyRyZXN1bHQsIHRlcm1fc2l6ZSA8IDYwMCkgIyBub3RoaW5nIG92ZXItcmVwcmVzZW50ZWQsIGVtcHR5ISAKCm9yYV9yZXZfdmFyX3Byb3RzMiA8LSBnb3N0KCBxdWVyeSA9IHVuaXF1ZShyZXZfdmFyX3Byb3RzMiRtZ2lfc3ltYm9sKSwKICAgICAgICAgICAgICAgICAgICAgb3JnYW5pc20gPSAibW11c2N1bHVzIiwKICAgICAgICAgICAgICAgICAgICAgZG9tYWluX3Njb3BlID0gImN1c3RvbSIsCiAgICAgICAgICAgICAgICAgICAgIGN1c3RvbV9iZyA9IHNoYXJlZC5nZW5lcyRtZ2lfc3ltYm9sLAogICAgICAgICAgICAgICAgICAgICBldmNvZGVzID0gVFJVRSwKICBjb3JyZWN0aW9uX21ldGhvZCA9ICJmZHIiCiAgICAgICAgICAgICAgICAgICAgICkKb3JhX3Jldl92YXJfcHJvdHMyJHJlc3VsdCA8LSBmaWx0ZXIoIG9yYV9yZXZfdmFyX3Byb3RzMiRyZXN1bHQsIHRlcm1fc2l6ZSA8IDYwMCkKCgpvcmFfcmV2X3Zhcl9wcm90czIkcmVzdWx0ICU+JQogc2VsZWN0KAogICAgYERhdGEgc291cmNlYCA9IHNvdXJjZSwKICAgIGBUZXJtIElEYCA9IHRlcm1faWQsCiAgICBgVGVybSBOYW1lYCA9IHRlcm1fbmFtZSwgCiAgICBgVGVybSBzaXplYCA9IHRlcm1fc2l6ZSwgCiAgICBgIyBvZiBpbnRlcnNlY3RpbmcgcHJvdGVpbnNgID0gaW50ZXJzZWN0aW9uX3NpemUsCiAgICAgRkRSID0gcF92YWx1ZQogICAgKSAlPiUgCiAgbXV0YXRlX2lmKCBpcy5udW1lcmljLCBmb3JtYXRDLCBkaWdpdHMgPTIpICU+JSAKICBjcmVhdGVfZHQoKQoKYGBgCgoKPGJyPgoKIyMjIyBGaWd1cmUgUzNECgpgYGB7ciBGaWd1cmVTM0QsIGZpZy5jYXA9IkZpZ3VyZSBTM0Q6IEdlbmV0aWNhbGx5IGlkZW50aWNhbCBjZWxsIGxpbmVzIHNob3cgc2lnbmlmaWNhbnRseSBoaWdoZXIgY29ycmVsYXRpb24gdGhhbiB3aGF0IGlzIGV4cGVjdGVkIGJ5IGNoYW5jZSBiZXR3ZWVuIHRoZSB0cmFuc2NyaXB0b21lIGFuZCBwcm90ZW9tZS4gVmlvbGluIHBsb3RzIG92ZXJsYWlkIHdpdGggYm94cGxvdHMgZGVwaWN0aW5nIHRoZSBkaXN0cmlidXRpb24gb2YgUGVhcnNvbiBjb3JyZWxhdGlvbiBjb2VmZmljaWVudHMgYmV0d2VlbiB0aGUgdHJhbnNjcmlwdG9tZSBhbmQgcHJvdGVvbWUgb2YgZ2VuZXRpY2FsbHkgaWRlbnRpY2FsIG1FU0NzIChibHVlKSBhbmQgdGhlIG51bGwgZGlzdHJpYnV0aW9uIGdlbmVyYXRlZCB0aHJvdWdoIDEwMDAgcGVybXV0YXRpb25zIHdoZXJlIHRoZSBzYW1wbGUgbmFtZXMgYXJlIHJhbmRvbWl6ZWQgKGJsYWNrKS4iLCB3YXJuaW5nPUZBTFNFLCBtZXNzYWdlPUZBTFNFLCBmaWcuaGVpZ2h0PTUsIGZpZy53aWR0aD00fQoKCiMgVGhlIGNvZGUgYmVsb3cgaXMgdXNlZCB0byBnZW5lcmF0ZSB0aGUgbnVsbCBkaXN0cmlidXRpb24uCiMgSXQgaXMgY29tbWVudGVkIG91dCBhbmQgbG9hZGVkIGZyb20gdGhlIHNhdmVkIFJkYXRhIGZpbGUgYmVjYXVzZSBpdCB0YWtlcyB0b28gbG9uZyB0byBydW4gd2l0aGluIHRoZSBzY3JpcHQuIAojIHNhbXBsZV9jb3IgPC0gYygpCiMgZm9yKCBpIGluIDE6MTAwMCl7CiMgICAjIHJhbmRvbWl6aW5nIHRoZSBzYW1wbGUgbmFtZXMgMTAwMCB0aW1lcyBhbmQgZ2V0dGluZyBjb3JyZWxhdGlvbnMKIyAKIyAgIHNoYXJlZF9wcm90X21hdCA8LSAgdChleHByWi5lc2NfcHJvdFtzaGFyZWQuc2FtcGxlcywgc2hhcmVkLnByb3QubmFtZXMkcHJvdGVpbl9pZF0pCiMgICAjIHJhbmRvbWl6ZSB0aGUgc2FtcGxlIG5hbWVzCiMgICBjb2xuYW1lcyhzaGFyZWRfcHJvdF9tYXQpIDwtIHBhc3RlMCggc2FtcGxlKGNvbG5hbWVzKHNoYXJlZF9wcm90X21hdCksIG5jb2woc2hhcmVkX3Byb3RfbWF0KSksIl9wcm90ZWluIikKIyAKIyAgIHNoYXJlZF9ybmFfbWF0IDwtICB0KGV4cHJaLmVzY19ybmFbc2hhcmVkLnNhbXBsZXMsIHNoYXJlZC5wcm90Lm5hbWVzJGVuc2VtYmxfZ2VuZV9pZF0pCiMgICAjIHJhbmRvbWl6ZSB0aGUgc2FtcGxlIG5hbWVzCiMgICBjb2xuYW1lcyhzaGFyZWRfcm5hX21hdCkgPC0gcGFzdGUwKCBzYW1wbGUoY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpLCBuY29sKHNoYXJlZF9ybmFfbWF0KSksIl9ybmEiKQojIAojICAgbWVhc3VyZS5jb3IuZGYgPC0gcmNvcnIoIHggPSBzaGFyZWRfcHJvdF9tYXQsCiMgICAgICAgICAgICAgICAgICAgICAgICAgIHkgPSBzaGFyZWRfcm5hX21hdCwKIyAgICAgICAgICAgICAgICAgICAgICAgICAgdHlwZSA9ICJwZWFyc29uIikKIyAKIyAgIHNhbXBsZV9jb3JbW2ldXSA8LSBhc190aWJibGUoIG1lYXN1cmUuY29yLmRmJHJbY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0KSwgY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpXSwKIyAgICAgICAgICAgICAgICAgICAgICAgICAgICAgcm93bmFtZXMgPSAicHJvdGVpbl9zYW1wbGUiKSAlPiUKIyAgIHBpdm90X2xvbmdlciggY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpLCBuYW1lc190byA9ICJybmFfc2FtcGxlIiwgdmFsdWVzX3RvID0gInIiKSAlPiUKIyAgIGlubmVyX2pvaW4oIChhc190aWJibGUoIG1lYXN1cmUuY29yLmRmJFBbY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0KSwgY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQpXSwKIyAgICAgICAgICAgICAgICAgICAgICAgICAgICAgcm93bmFtZXMgPSAicHJvdGVpbl9zYW1wbGUiKSAlPiUKIyAgICAgICBwaXZvdF9sb25nZXIoIGNvbG5hbWVzKHNoYXJlZF9ybmFfbWF0KSwgbmFtZXNfdG8gPSAicm5hX3NhbXBsZSIsIHZhbHVlc190byA9ICJwX3ZhbCIpICkgKSAlPiUKIyAgICAgbXV0YXRlKCBuID0gaSkKIyAKIyB9CiMgCiMgc2F2ZShzYW1wbGVfY29yLCBmaWxlID0gaGVyZSgiX2RhdGEiLCJybmFfcHJvdF9zYW1wbGVfY29yX3Blcm1fcGVhcnNvbi5SRGF0YSIpKQoKCmxvYWQoaGVyZSgiLi4vcFFUTF93ZWJzaXRlL19kYXRhIiwicm5hX3Byb3Rfc2FtcGxlX2Nvcl9wZXJtX3BlYXJzb24uUkRhdGEiKSkKCnNhbXBsZV9jb3IgJT4lIAogIGVuZnJhbWUoKSAlPiUgCiAgdW5uZXN0KHZhbHVlKSAlPiUgCiAgbXV0YXRlKCBwcm90ZWluX3NhbXBsZSA9IGdzdWIoIl9wcm90ZWluIiwiIixwcm90ZWluX3NhbXBsZSkgLAogICAgICAgICAgcm5hX3NhbXBsZSA9IGdzdWIoIl9ybmEiLCIiLHJuYV9zYW1wbGUpKSAlPiUgCiAgZmlsdGVyKCBwcm90ZWluX3NhbXBsZSA9PSBybmFfc2FtcGxlKSAtPiBudWxsX3NhbXBsZV9jb3JfZGlzdAoKcHJvdF9ybmFfc2FtcGxlX2Nvcl9kZiAlPiUKICBtdXRhdGUoIHNhbXBsZWlkX3Byb3QgPSBnc3ViKCJfcHJvdGVpbiIsIiIscHJvdGVpbl9zYW1wbGUpLAogICAgICAgICAgc2FtcGxlaWRfcm5hID0gZ3N1YigiX3JuYSIsIiIsIHJuYV9zYW1wbGUpKSAlPiUKICBmaWx0ZXIoIHNhbXBsZWlkX3JuYSA9PSBzYW1wbGVpZF9wcm90KSAtPiByZWFsX3NhbXBsZV9jb3JfZGlzdAoKbnVsbF9zYW1wbGVfY29yX2Rpc3QgJT4lIAogIG11dGF0ZSggdHlwZSA9ICJOdWxsIikgJT4lIAogIHNlbGVjdCggdHlwZSwgcikgJT4lIAogIHJiaW5kKCByZWFsX3NhbXBsZV9jb3JfZGlzdCAlPiUgCiAgICAgICAgICAgbXV0YXRlKCB0eXBlID0gIlJlYWwiKSAlPiUgCiAgICAgICAgICAgc2VsZWN0KCB0eXBlLCByKSkgJT4lIAogIGdncGxvdCgpKwogIGFlcyggeCA9IHR5cGUsCiAgICAgICB5ID0gciwgCiAgICAgICBjb2wgPSB0eXBlICkrCiAgZ2VvbV92aW9saW4oIHNob3cubGVnZW5kID0gRikrCiAgZ2VvbV9ib3hwbG90KHdpZHRoID0gMC4xLCBzaG93LmxlZ2VuZCA9IEYpKwogIHNjYWxlX2NvbG9yX21hbnVhbCggdmFsdWVzID0gYygiYmxhY2siLCJibHVlIikpKwogIHRoZW1lX3B1YmNsZWFuKGJhc2Vfc2l6ZSA9IDE4KSsKICB0aGVtZShsZWdlbmQucG9zaXRpb249Im5vbmUiKSsKICB5bGFiKCJDb3JyZWxhdGlvbiIpKwogIHhsYWIoIkRpc3RyaWJ1dGlvbiIpKwogIHN0YXRfY29tcGFyZV9tZWFucyggbGFiZWwueSA9IDAuNjIsIGxhYmVsLnggPSAxLjE1KSsKICB5bGltKC0wLjUsLjY1KSAtPiBmaWdfczNkCiAgCmdnYXJyYW5nZShmaWdfczNkLCAKICAgICAgICAgIGxhYmVscyA9ICJEIiwKICAgICAgICAgIGZvbnQubGFiZWwgPSBsaXN0KCBzaXplID0gMjApCiAgICAgICAgICAgICAgICAgICAgICApCgpgYGAKCgo8YnI+Cjxicj4KCiMjIyBGaWd1cmUgM0M6IENvdmFyaWF0aW9uIG9mIHByb3RlaW4gYW5kIHRyYW5zY3JpcHQgYWJ1bmRhbmNlIGFjcm9zcyBnZW5lcwoKYGBge3IgRmlndXJlM0NfcHJlcCwgY2FjaGUgPSBUUlVFLCBjYWNoZS5sYXp5PUZBTFNFfQoKIyBnZXQgZ2VuZSBuYW1lcyBmb3IgYWxsIHByb3RlaW4gaWRzCnNoYXJlZC5wcm90Lm5hbWVzIDwtIHNoYXJlZC5nZW5lcyAlPiUKICBncm91cF9ieShlbnNlbWJsX2dlbmVfaWQpICU+JQogIG11dGF0ZShuZXdfc3ltYm9sID0gcGFzdGUwKG1naV9zeW1ib2wsICJfIiwgMTpuKCkpLAogICAgICAgICBuZXdfZ2VuZV9pZCA9IHBhc3RlMChlbnNlbWJsX2dlbmVfaWQsICJfIiwgMTpuKCkpKQoKIyBjaGFuZ2UgaWQncyBpbnRvIGdlbmUgc3ltYm9scwpzaGFyZWRfcHJvdF9tYXQyIDwtICAoZXhwclouZXNjX3Byb3Rbc2hhcmVkLnNhbXBsZXMsIHNoYXJlZC5wcm90Lm5hbWVzJHByb3RlaW5faWRdKQpzaGFyZWRfcm5hX21hdDIgPC0gIChleHByWi5lc2Nfcm5hW3NoYXJlZC5zYW1wbGVzLCBzaGFyZWQucHJvdC5uYW1lcyRlbnNlbWJsX2dlbmVfaWRdKQoKIyBnZXQgZ2VuZSBsZXZlbCBjb3JyZWxhdGlvbnMKcHJvdF9ybmFfZ2VuZV9jb3IgPC0gcmNvcnIoIHggPSBzaGFyZWRfcHJvdF9tYXQyLAogICAgICAgICAgICAgICAgICAgICAgICAgeSA9IHNoYXJlZF9ybmFfbWF0MiwKICAgICAgICAgICAgICAgICAgICAgICAgIHR5cGUgPSAicGVhcnNvbiIpCgojIGNvbnZlcnQgZ2VuZSBsZXZlbCBjb3JyZWxhdGlvbnMgdG8gZGF0YSBmcmFtZSAKcHJvdF9ybmFfZ2VuZV9jb3JfZGYgPC0gdGliYmxlKCByID1kaWFnKHByb3Rfcm5hX2dlbmVfY29yJHJbY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0MiksIGNvbG5hbWVzKHNoYXJlZF9ybmFfbWF0MildKSwKICAgICAgICAgICAgICAgICAgICAgICBwX3ZhbCA9IGRpYWcocHJvdF9ybmFfZ2VuZV9jb3IkUFtjb2xuYW1lcyhzaGFyZWRfcHJvdF9tYXQyKSwgY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQyKV0pLAogICAgICAgICAgICAgICAgICAgICAgIG4gPSBkaWFnKHByb3Rfcm5hX2dlbmVfY29yJG5bY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0MiksIGNvbG5hbWVzKHNoYXJlZF9ybmFfbWF0MildKSwKICAgICAgICAgICAgICAgICAgICAgICBwcm90ZWluX2lkID0gY29sbmFtZXMoc2hhcmVkX3Byb3RfbWF0MiksCiAgICAgICAgICAgICAgICAgICAgICAgZW5zZW1ibF9nZW5lX2lkID0gY29sbmFtZXMoc2hhcmVkX3JuYV9tYXQyKQogICAgICAgICAgICAgICAgICAgICAgICkgJT4lCiAgbGVmdF9qb2luKC4sIHNoYXJlZC5wcm90Lm5hbWVzKSAlPiUKICBtdXRhdGUocF9hZGogPSBwLmFkanVzdChwX3ZhbCwgbWV0aG9kID0gIkJIIikpICU+JQogIG11dGF0ZSggY29yID0gcikgJT4lCiAgYXJyYW5nZSgoZGVzYyhjb3IpKSkgJT4lCiAgbXV0YXRlKHJhbmsgPSBzZXEoMTpuKCkpKQoKIyBnZXQgc2lnbmlmaWNhbnRseSBuZWdhdGl2ZWx5IGFuZCBwb3NpdGl2ZWx5IGNvcnJlbGF0ZWQgZ2VuZXMgYW5kIGdlbmVzIHdpdGggbm90IGNvcnJlbGF0aW9uIChhYnMoY29yKSA8MC4wNSkuCm5lZ19jb3IgPC0gcHJvdF9ybmFfZ2VuZV9jb3JfZGYgJT4lCiAgZmlsdGVyKCBjb3IgPCAwLCBwX2FkaiA8IDAuMDUpCgpwb3NfY29yIDwtIHByb3Rfcm5hX2dlbmVfY29yX2RmICU+JQogIGZpbHRlciggY29yID4gMCwgcF9hZGogPCAwLjA1KSAlPiUKICBhcnJhbmdlKCBkZXNjKGNvcikgKQoKbm9fY29yIDwtIHByb3Rfcm5hX2dlbmVfY29yX2RmICU+JQogIGZpbHRlciggYWJzKGNvcikgPCAwLjA1KSAKCiMgb3Zlci1yZXByZXNlbnRhdGlvbiBhbmFseXNpcyB3aXRoIGVhY2ggY2F0ZWdvcnkKb3JhX25lZ19jb3IgPC0gZ29zdCggcXVlcnkgPSBuZWdfY29yJG1naV9zeW1ib2wsCiAgICAgICAgICAgICAgICAgICAgIG9yZ2FuaXNtID0gIm1tdXNjdWx1cyIsCiAgICAgICAgICAgICAgICAgICAgIGRvbWFpbl9zY29wZSA9ICJjdXN0b20iLAogICAgICAgICAgICAgICAgICAgICBjdXN0b21fYmcgPSBwcm90X3JuYV9nZW5lX2Nvcl9kZiRtZ2lfc3ltYm9sLAogICAgICAgICAgICAgICAgICAgICBldmNvZGVzID0gVFJVRSwKICBjb3JyZWN0aW9uX21ldGhvZCA9ICJmZHIiCiAgICAgICAgICAgICAgICAgICAgICkKb3JhX25lZ19jb3IkcmVzdWx0IDwtIGZpbHRlciggb3JhX25lZ19jb3IkcmVzdWx0LCB0ZXJtX3NpemUgPCA2MDApCgpvcmFfcG9zX2NvciA8LSBnb3N0KCBxdWVyeSA9IHBvc19jb3IkbWdpX3N5bWJvbCwKICAgICAgICAgICAgICAgICAgICAgb3JnYW5pc20gPSAibW11c2N1bHVzIiwKICAgICAgICAgICAgICAgICAgICAgZG9tYWluX3Njb3BlID0gImN1c3RvbSIsCiAgICAgICAgICAgICAgICAgICAgIGN1c3RvbV9iZyA9IHByb3Rfcm5hX2dlbmVfY29yX2RmJG1naV9zeW1ib2wsCiAgICAgICAgICAgICAgICAgICAgIGV2Y29kZXMgPSBUUlVFLAogIGNvcnJlY3Rpb25fbWV0aG9kID0gImZkciIKICAgICAgICAgICAgICAgICAgICAgKQpvcmFfcG9zX2NvciRyZXN1bHQgPC0gZmlsdGVyKCBvcmFfcG9zX2NvciRyZXN1bHQsIHRlcm1fc2l6ZSA8IDYwMCkKCm9yYV9ub19jb3IgPC0gZ29zdCggcXVlcnkgPSBub19jb3IkbWdpX3N5bWJvbCwKICAgICAgICAgICAgICAgICAgICAgb3JnYW5pc20gPSAibW11c2N1bHVzIiwKICAgICAgICAgICAgICAgICAgICAgZG9tYWluX3Njb3BlID0gImN1c3RvbSIsCiAgICAgICAgICAgICAgICAgICAgIGN1c3RvbV9iZyA9IHByb3Rfcm5hX2dlbmVfY29yX2RmJG1naV9zeW1ib2wsCiAgICAgICAgICAgICAgICAgICAgIGV2Y29kZXMgPSBUUlVFLAogIGNvcnJlY3Rpb25fbWV0aG9kID0gImZkciIKICAgICAgICAgICAgICAgICAgICAgKQpvcmFfbm9fY29yJHJlc3VsdCA8LSBmaWx0ZXIoIG9yYV9ub19jb3IkcmVzdWx0LCB0ZXJtX3NpemUgPCA2MDApCgojIG1lcmdlIGFsbCBPUkEgcmVzdWx0cyBpbnRvIG9uZSBkYXRhIGZyYW1lCm9yYV9uZWdfY29yJHJlc3VsdCAlPiUKICBtdXRhdGUoIGdyb3VwID0gIk5lZ2F0aXZlIGNvcnJlbGF0aW9uIikgJT4lCiAgcmJpbmQoIG11dGF0ZSggb3JhX3Bvc19jb3IkcmVzdWx0LCBncm91cCA9ICJQb3NpdGl2ZSBjb3JyZWxhdGlvbiIpKSAlPiUKICByYmluZCggbXV0YXRlKCBvcmFfbm9fY29yJHJlc3VsdCAsIGdyb3VwID0gIk5vIGNvcnJlbGF0aW9uIikpIC0+IG9yYV9hbGxfY29ycgoKCkZpZ3VyZTNDX2RhdGFfcGFydDEgPC0gcHJvdF9ybmFfZ2VuZV9jb3JfZGYgJT4lIAogIG11dGF0ZSggCiAgICBHcm91cCA9IGNhc2Vfd2hlbigKICAgICAgKGNvciA8IDAgJiBwX2FkaiA8IDAuMDUpIH4gIk5lZ2F0aXZlIiwKICAgICAgKGNvciA+IDAgJiBwX2FkaiA8IDAuMDUpIH4gIlBvc2l0aXZlIiwKICAgICAgKCBhYnMoY29yKSA8IDAuMDUpIH4gIkxvdyIKICAgICkKICAgICkgJT4lIAogICBzZWxlY3QoIAogICAgYEdlbmUgbmFtZWAgPSBtZ2lfc3ltYm9sLAogICAgYFByb3RlaW4gSURgID0gcHJvdGVpbl9pZCwKICAgIGBHZW5lIElEYCA9IGVuc2VtYmxfZ2VuZV9pZCwgCiAgICBgQ29ycmVsYXRpb25gID0gY29yLAogICAgR3JvdXAKICAgICkKCiMgZ2V0IHRoZXNlOgojIG5lZ2F0aXZlOiBjZWxsdWxhciByZXNwaXJhdGlvbiwgbWl0b2Nob25kcmlhbCByaWJvc29tZQojIG5vIGNvcnJlbGF0aW9uOiBSaWJvc29tZSwgU3BsaWNlb3NvbWUgT1IgIGN5dG9wbGFzbWljIHRyYW5zbGF0aW9uLCBtUk5BIHNwbGljaW5nLCB2aWEgc3BsaWNlb3NvbWUKIyBwb3NpdGl2ZSBjb3JyZWxhdGlvbjogZXh0cmFjZWxsdWxhciByZWdpb24sIGxpcGlkIG1ldGFib2xpYyBwcm9jZXNzCiMgcmFuayBoZXJlIGlzIHRoZSBvcmRlciBhYm92ZQojIEZvciBhZGRpbmcgdGhlIGRvdHMgYmVsb3cgdGhlIGNvcnJlbGF0aW9uIGhpc3RvZ3JhbQpGaWd1cmUzQ19kYXRhX3BhcnQyIDwtIG9yYV9hbGxfY29yciAlPiUKICAjZmlsdGVyKHNvdXJjZSAlaW4lIGMoIkdPOkJQIiwiUkVBQyIsIktFR0ciKSkgJT4lCiAgZmlsdGVyKHRlcm1fbmFtZSAlaW4lIGMoImNlbGx1bGFyIHJlc3BpcmF0aW9uIiwgIm1pdG9jaG9uZHJpYWwgcmlib3NvbWUiLAogICAgICAgICAgICAgICAgICAgICAgICAgICJjeXRvcGxhc21pYyB0cmFuc2xhdGlvbiIsICJtUk5BIHNwbGljaW5nLCB2aWEgc3BsaWNlb3NvbWUiLAogICAgICAgICAgICAgICAgICAgICAgICAgICJleHRyYWNlbGx1bGFyIHJlZ2lvbiIsICJsaXBpZCBtZXRhYm9saWMgcHJvY2VzcyIsIlgtbGlua2VkIGluaGVyaXRhbmNlIikgKSAlPiUKICBzZWxlY3QodGVybV9uYW1lLCBpbnRlcnNlY3Rpb24sIHBfdmFsdWUsIGdyb3VwKSAlPiUKICBhcnJhbmdlKGRlc2MocF92YWx1ZSksIGdyb3VwKSAlPiUKICBzZXBhcmF0ZV9yb3dzKGludGVyc2VjdGlvbikgJT4lCiAgbGVmdF9qb2luKC4sIHByb3Rfcm5hX2dlbmVfY29yX2RmLCBieSA9IGMoImludGVyc2VjdGlvbiIgPSAibWdpX3N5bWJvbCIpKSAlPiUgCiAgbXV0YXRlKCBncm91cDIgPSBjYXNlX3doZW4oICAgKCBjb3IgPCAwICYgcF9hZGogPCAwLjA1KSB+ICJuZWdhdGl2ZSIsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAoIGNvciA+IDAgJiBwX2FkaiA8IDAuMDUpIH4gInBvc2l0aXZlIiwKICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICggYWJzKGNvcikgPD0gMC4wNSApIH4gImxvdyIsCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAoIGFicyhjb3IpID4gMC4wNSAmIHBfYWRqID49IDAuMDUpIH4gIm5vbmUiCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICkKICApCnlfY29sb3JzIDwtIFJDb2xvckJyZXdlcjo6YnJld2VyLnBhbChuID0gNCwgbmFtZSA9ICJEYXJrMiIpCgojIEZvciBhZGRpbmcgdGhlIGxhYmVscyBuZXh0IHRvIGRvdHMgaW4gbWF0Y2hpbmcgY29sb3IKRmlndXJlM0NfZGF0YV9wYXJ0MyA8LSBvcmFfYWxsX2NvcnIgJT4lCiAgZmlsdGVyKHRlcm1fbmFtZSAlaW4lIGMoImNlbGx1bGFyIHJlc3BpcmF0aW9uIiwgIm1pdG9jaG9uZHJpYWwgcmlib3NvbWUiLAogICAgICAgICAgICAgICAgICAgICAgICAgICJjeXRvcGxhc21pYyB0cmFuc2xhdGlvbiIsICJtUk5BIHNwbGljaW5nLCB2aWEgc3BsaWNlb3NvbWUiLAogICAgICAgICAgICAgICAgICAgICAgICAgICJleHRyYWNlbGx1bGFyIHJlZ2lvbiIsICJsaXBpZCBtZXRhYm9saWMgcHJvY2VzcyIsIlgtbGlua2VkIGluaGVyaXRhbmNlIikgKSAlPiUKICBzZWxlY3QodGVybV9uYW1lLCBpbnRlcnNlY3Rpb24sIHBfdmFsdWUsIGdyb3VwKSAlPiUKICBtdXRhdGUodGVybV9uYW1lID0gcGFzdGUodGVybV9uYW1lLCBmb3JtYXRDKHBfdmFsdWUsIGZvcm1hdCA9ICJlIiwgZGlnaXRzID0gMikpKSAlPiUKICBzZXBhcmF0ZV9yb3dzKGludGVyc2VjdGlvbikgJT4lCiAgbGVmdF9qb2luKC4sIHByb3Rfcm5hX2dlbmVfY29yX2RmLCBieSA9IGMoImludGVyc2VjdGlvbiIgPSAibWdpX3N5bWJvbCIpKSAlPiUKICBncm91cF9ieSh0ZXJtX25hbWUsIGdyb3VwLCBwX3ZhbHVlKSAlPiUKICBkcGx5cjo6c3VtbWFyaXplKG1lZF9jb3IgPSBtZWRpYW4oY29yLCBuYS5ybSA9IFRSVUUpKSAlPiUKICBtdXRhdGUoeV9jb2wgPSBpZmVsc2UoZ3JvdXAgPT0gIk5lZ2F0aXZlIGNvcnJlbGF0aW9uIiwgeV9jb2xvcnNbMl0seV9jb2xvcnNbNF0pLAogICAgICAgICB5X2NvbCA9IGlmZWxzZShncm91cCA9PSAiTm8gY29ycmVsYXRpb24iLCB5X2NvbG9yc1szXSwgeV9jb2wpLAogICAgICAgICB5X2NvbCA9IGlmZWxzZShncm91cCA9PSAiUG9zaXRpdmUgY29ycmVsYXRpb24iLCB5X2NvbG9yc1sxXSwgeV9jb2wpKSAlPiUKICBhcnJhbmdlKGdyb3VwLCBwX3ZhbHVlKSAKCgpgYGAKCgpgYGB7ciBGaWd1cmUzQywgZmlnLmNhcCA9ICJGaWd1cmUgM0M6IEdlbmVzIHNob3cgdmFyaWFibGUgYWdyZWVtZW50IGJldHdlZW4gdHJhbnNjcmlwdCBhbmQgcHJvdGVpbiBhYnVuZGFuY2UgaW4gRE8gbUVTQ3MuIEhpc3RvZ3JhbSBkZXBpY3RpbmcgdGhlIGRpc3RyaWJ1dGlvbiBvZiBwYWlyd2lzZSBQZWFyc29uIGNvcnJlbGF0aW9uIGNvZWZmaWNpZW50cyBiZXR3ZWVuIHRyYW5zY3JpcHQgYW5kIHByb3RlaW4gYWJ1bmRhbmNlIG9mIGdlbmVzIHdpdGggY2hhcmFjdGVyaXN0aWMgR08gdGVybXMgb3ZlcnJlcHJlc2VudGVkIGluIGVhY2ggY2F0ZWdvcnkgYW5ub3RhdGVkIHVuZGVybmVhdGggaW4gbWF0Y2hpbmcgY29sb3JzIChncmVlbjogc2lnbmlmaWNhbnRseSBwb3NpdGl2ZWx5IGNvcnJlbGF0ZWQsIG9yYW5nZTogc2lnbmlmaWNhbnRseSBuZWdhdGl2ZWx5IGNvcnJlbGF0ZWQsIHB1cnBsZTogZ2VuZXMgd2l0aCBsaXR0bGUgb3Igbm8gY29ycmVsYXRpb24pLiIsIHdhcm5pbmc9RkFMU0UsIG1lc3NhZ2U9RkFMU0UsIGZpZy53aWR0aD0xMCwgZmlnLmhlaWdodD02fQoKCgojIHBsb3QgcmVzdWx0cwp5X2NvbG9ycyA8LSBSQ29sb3JCcmV3ZXI6OmJyZXdlci5wYWwobiA9IDQsIG5hbWUgPSAiRGFyazIiKQoKRmlndXJlM0NfZGF0YV9wYXJ0MSAlPiUgCiAgZ2dwbG90KCkgKwogIGFlcyh4ID0gQ29ycmVsYXRpb24pICsKICBnZW9tX2hpc3RvZ3JhbShhZXMoZmlsbCA9IEdyb3VwKSwgc2hvdy5sZWdlbmQgPSBGLCBiaW53aWR0aCA9IDAuMDEsIGFscGhhID0gMC43KSArCiAgeGxhYigiR2VuZSBjb3JyZWxhdGlvbiIpICsKICB5bGFiKCIiKSsKICBzY2FsZV9maWxsX21hbnVhbChsaW1pdHMgPSBjKCAiTG93IixOQSwgIk5lZ2F0aXZlIiwgIlBvc2l0aXZlIiksIHZhbHVlcyA9IGMoeV9jb2xvcnNbM10sImdyYXkiLHlfY29sb3JzWzJdLHlfY29sb3JzWzFdKSApICsKICB0aGVtZV9wdWJjbGVhbihiYXNlX3NpemUgPSAxOCwgYmFzZV9mYW1pbHkgPSAiUG9wcGlucyIpKwogIGdlb21fdmxpbmUoIGFlcyh4aW50ZXJjZXB0ID0gbWVkaWFuKENvcnJlbGF0aW9uKSksIGxpbmV0eXBlID0gMiwgY29sID0gImJsYWNrIiwgc2l6ZSA9IDEpICsKICBhbm5vdGF0ZSgidGV4dCIsCiAgICB4ID0gbWVkaWFuKEZpZ3VyZTNDX2RhdGFfcGFydDEkQ29ycmVsYXRpb24sIG5hLnJtID0gVCkgLTAuMjUsIHkgPSAxMzUsCiAgICBsYWJlbCA9IHBhc3RlMCgiTWVkaWFuID0gXG4iLCBmb3JtYXRDKG1lZGlhbihGaWd1cmUzQ19kYXRhX3BhcnQxJENvcnJlbGF0aW9uLCBuYS5ybSA9IFQpLCBkaWdpdHMgPSAxLCBmb3JtYXQgPSJmIikpLAogICAgc2l6ZSA9IDYsCiAgICBmYW1pbHkgPSAiUG9wcGlucyIKICApKwogIHhsaW0oLTAuNSwgMSkrCiAgeWxpbSgwLDE1MCktPiBwMQoKCkZpZ3VyZTNDX2RhdGFfcGFydDIgJT4lIAogIGFycmFuZ2UoY29yKSAlPiUgCiAgbXV0YXRlKGxhYmVsMiA9IGZhY3RvcihwYXN0ZSh0ZXJtX25hbWUsIGZvcm1hdEMocF92YWx1ZSwgZm9ybWF0ID0gImUiLCBkaWdpdHMgPSAyKSksCiAgICBsZXZlbHMgPSBGaWd1cmUzQ19kYXRhX3BhcnQzJHRlcm1fbmFtZQogICkpICU+JQogIG11dGF0ZShsYWJlbCA9IGZhY3RvcihpbnRlcnNlY3Rpb24sIGxldmVscyA9IHVuaXF1ZShpbnRlcnNlY3Rpb24pKSkgJT4lCiAgZ2dwbG90KCkgKwogIGFlcyh5ID0gbGFiZWwyLCB4ID0gY29yLCBjb2wgPSBncm91cDIsIGdyb3VwID0gdGVybV9uYW1lKSArCiAgZ2VvbV9wb2ludChzaG93LmxlZ2VuZCA9IEZBTFNFLCBzaGFwZSA9IDE1KSArCiAgc2NhbGVfY29sb3JfbWFudWFsKGxpbWl0cyA9IGMoImxvdyIsICJub25lIiwgIm5lZ2F0aXZlIiwgInBvc2l0aXZlIiksIHZhbHVlcyA9IGMoeV9jb2xvcnNbM10sImdyYXkiLHlfY29sb3JzWzJdLHlfY29sb3JzWzFdKSApICsKICB4bGFiKCJHZW5lIGNvcnJlbGF0aW9uIikgKwogIHlsYWIoIiIpICsKICB0aGVtZV9wdWJjbGVhbihiYXNlX3NpemUgPSAyMCwgYmFzZV9mYW1pbHkgPSAiUG9wcGlucyIpICsKICB0aGVtZSgKICAgIGF4aXMudGl0bGUueCA9IGVsZW1lbnRfYmxhbmsoKSwKICAgIGF4aXMudGV4dC54ID0gZWxlbWVudF9ibGFuaygpLAogICAgYXhpcy50aWNrcy54ID0gZWxlbWVudF9ibGFuaygpLAogICAgYXhpcy50ZXh0LnkgPSBlbGVtZW50X3RleHQoc2l6ZSA9IDEzLCBjb2xvciA9IChGaWd1cmUzQ19kYXRhX3BhcnQzJHlfY29sKSkKICApICsKICBzY2FsZV95X2Rpc2NyZXRlKHBvc2l0aW9uID0gInJpZ2h0IikgKwogIHhsaW0oLTAuNSwgMSktPiBwMgoKdG9wX3JvdyA8LSBwbG90X2dyaWQocDEsIE5VTEwsIHJlbF93aWR0aHMgPSBjKDEsIDAuNykpCmJvdHRvbV9yb3cgPC0gcGxvdF9ncmlkKE5VTEwsIHAyLCByZWxfd2lkdGhzID0gYygwLjA3LCAxKSkKb3JhX2Nvcl9wbG90PC0gcGxvdF9ncmlkKHRvcF9yb3csIGJvdHRvbV9yb3csIG5yb3cgPSAyLCByZWxfaGVpZ2h0cyA9IGMoMSwgMSkpIAoKb3JhX2Nvcl9wbG90CgpgYGAKCjxicj4KCmBgYHtyIEZpZ3VyZTNDX2RhdGEsIHdhcm5pbmc9RkFMU0UsIG1lc3NhZ2U9RkFMU0UsIGZpZy5jYXAgPSJDb3JyZWxhdGlvbiB2YWx1ZXMgdXNlZCBpbiBwbG90dGluZyBGaWd1cmUgM0MgdG9wIHBhbmVsIHdpdGggZ3JvdXBzIGNvbG9yZWQuIn0KCkZpZ3VyZTNDX2RhdGFfcGFydDEgJT4lCiAgbXV0YXRlX2lmKCBpcy5udW1lcmljLCByb3VuZCwgMikgJT4lIAogIGNyZWF0ZV9kdCgpCiAgCgpgYGAKCjxicj4KCmBgYHtyIE9SQV9yZXN1bHRzLCB3YXJuaW5nPUZBTFNFLCBtZXNzYWdlPUZBTFNFLCBmaWcuY2FwPSJMaXN0IG9mIG92ZXItcmVwcmVzZW50ZWQgYmlvbG9naWNhbCBwcm9jZXNzZXMgYW5kIHBhdGh3YXlzIGluIGdlbmVzIHdpdGggcG9zaXRpdmUsIG5lZ2F0aXZlIGFuZCBubyBjb3JyZWxhdGlvbiBiZXR3ZWVuIHRyYW5zY3JpcHQgYW5kIHByb3RlaW4gYWJ1bmRhbmNlLiJ9CgoKb3JhX2FsbF9jb3JyICU+JQogc2VsZWN0KAogICBgR3JvdXBgPWdyb3VwLCAKICAgYERhdGEgc291cmNlYCA9IHNvdXJjZSwKICAgIGBUZXJtIElEYCA9IHRlcm1faWQsCiAgICBgVGVybSBOYW1lYCA9IHRlcm1fbmFtZSwgCiAgICBgVGVybSBzaXplYCA9IHRlcm1fc2l6ZSwgCiAgICBgIyBvZiBpbnRlcnNlY3RpbmcgcHJvdGVpbnNgID0gaW50ZXJzZWN0aW9uX3NpemUsCiAgICAgRkRSID0gcF92YWx1ZQogICAgKSAlPiUgCiAgbXV0YXRlX2lmKCBpcy5udW1lcmljLCBmb3JtYXRDLCBkaWdpdHMgPTIpICU+JSAKICBjcmVhdGVfZHQoKQoKYGBgCgoKPGJyPgoKIyMjIyBGaWd1cmUgUzNFIAoKYGBge3IgRmlndXJlUzNFLCBmaWcuY2FwID0gIkZpZ3VyZSBTM0U6IEdlbmVzIHRoYXQgZG8gbm90IGZvcm0gY29tcGxleGVzIHNob3cgc2lnbmlmaWNhbnRseSBoaWdoZXIgY29ycmVsYXRpb24gYmV0d2VlbiB0cmFuc2NyaXB0IGFuZCBwcm90ZWluIGFidW5kYW5jZS4gQm94cGxvdCBjb21wYXJpbmcgdGhlIHBhaXJ3aXNlIFBlYXJzb24gY29ycmVsYXRpb24gY29lZmZpY2llbnRzIGJldHdlZW4gdHJhbnNjcmlwdCBhbmQgcHJvdGVpbiBhYnVuZGFuY2UgZm9yIGdlbmVzIHRoYXQgYXJlIHBhcnQgb2YgcHJvdGVpbiBjb21wbGV4ZXMgKFRSVUUpIGFuZCB0aGF0IGRvIG5vdCBmb3JtIGNvbXBsZXhlcyAoRkFMU0UpLiIsIHdhcm5pbmc9RkFMU0UsIG1lc3NhZ2U9RkFMU0UsIGZpZy5oZWlnaHQ9NSwgZmlnLndpZHRoPTR9Cgpwcm90X3JuYV9nZW5lX2Nvcl9kZiAlPiUKICBtdXRhdGUoIHR5cGUgPSBpZmVsc2UoIGVuc2VtYmxfZ2VuZV9pZCAlaW4lIGNvbXBsZXguZ2VuZS5saXN0JGVuc2VtYmxfZ2VuZV9pZCwKICAgICAgICAgICAgICAgICAgICAgICAgIFRSVUUsCiAgICAgICAgICAgICAgICAgICAgICAgICBGQUxTRSkKICAgICAgICAgICkgJT4lCiAgZ2dwbG90KCkgKwogIGFlcyh5ID0gY29yLCB4ID0gdHlwZSkgKwogIGdlb21fYm94cGxvdCggd2lkdGggPSAwLjI1LCBhZXMoIGNvbCA9IHR5cGUpLCBzaG93LmxlZ2VuZCA9IEYpKwogICNnZW9tX2RlbnNpdHkoYWVzKGNvbCA9IHR5cGUsIGZpbGwgPSB0eXBlKSwgc2hvdy5sZWdlbmQgPSBULCBiaW5zID0gNDAwLCBhbHBoYSA9IDAuNikgKwogIHlsYWIoIkNvcmVsYXRpb24iKSArCiAgc2NhbGVfY29sb3JfbWFudWFsKHZhbHVlcyA9IGMoImdyYXkiLCJyZWQiKSkgKwogICNnZ3RpdGxlKCJUcmFuc2NyaXB0IHZzIFByb3RlaW4gYWJ1bmRhbmNlIGZvciBnZW5lcyIpICsKICB0aGVtZV9wdWJjbGVhbihiYXNlX3NpemUgPSAxOCkrCiAgeGxhYigiQ29tcGxleCBmb3JtaW5nIikrCiAgc3RhdF9jb21wYXJlX21lYW5zKCAgbGFiZWwueCA9IDEuMiwgbGFiZWwueSA9IDEuMSkrCiAgeWxpbSggLS41LCAxLjIpIC0+IGZpZ3VyZV9zM2UKCgpnZ2FycmFuZ2UoZmlndXJlX3MzZSwgICAgICAgICAgCiAgICAgICAgICBsYWJlbHMgPSAiRSIsCiAgICAgICAgICBmb250LmxhYmVsID0gbGlzdCggc2l6ZSA9IDIwKQogICAgICAgICAgICAgICAgICAgICAgKQoKYGBgCgoKCgoKCg==